BSDB Gurdon Studentship Report – Paula Richter
Posted by Paula Richter, on 10 January 2023
NRAP H100Q Mutation in Zebrafish
During the first year of my course in biological sciences, I became interested in developmental biology. I am mesmerized by the exploration of the beginnings of life through the complex interactions and changes that are taking place from the moment of fertilization. Furthermore, I realized the potential applications that scientific advancements in this area may have for disease prevention and treatment. This is especially interesting to me as a wildlife health student, aiming to conduct research on wildlife conservation in the future.
Research led by Dr. Denis Larkin found a mutation in the amino acid sequence (AA) of the NRAP gene in Yakut Cattle, resulting in a single nucleotide polymorphism (SNP resulting in a Glutamine to Histidine substitution (H100Q) at position 100) (Larkin et al., 2021). Yakut cattle are resistant to the low temperatures of the Siberian arctic and exhibit features such as efficient thermoregulation, a slow metabolic rate, and resistance to diseases. Nebulin-Related-Anchoring Protein (NRAP) is an actin-binding gene responsible for producing proteins to connect actin filaments in skeletal and cardiac muscle (GeneCards, 2022). The NRAP gene AA sequence is evolutionary conserved, however, this specific H100Q mutation was found in other cold-adapted, hibernating species and those entering periods of extreme bradycardia.
The expression of this particular SNP in those specific animal species prompted us to start this study. My summer project aimed to create the H100Q NRAP single-point mutation in the human gene and clone it into a suitable vector for in-vivo expression. This construct will then be used for microinjections into zebrafish embryos to observe potential changes in their heart development, heart rate, and muscle function.
I was keen to start the project and started by gathering information about zebrafish heart development, as well as molecular biology techniques. Together with my team, we created a well-thought-out cloning and mutation strategy to substitute a single nucleotide from the human NRAP gene from CAC (H) to CAA (Q) to recreate the H100Q mutation found in Yakut cattle.
However, I soon learned that theory and practice are two very different sides of the same coin. The world of molecular biology seemed to be unpredictable at first! After the first couple of ligations that didn’t produce any colonies, it was difficult to believe that getting enough material to mutate this gene was even possible. But that didn’t stop us. We troubleshot, experimented with enzymes and buffers, made our own competent cells and bacteria plates, changed reaction conditions, consulted papers, and of course, could always rely on the advice of our helpful supervisors. Having an amazing and experienced team around us at all times was key to our success. Not only could they provide valuable insights through their many years of experience in molecular biology but also offered constant encouragement: “it will be fine” turned out to be right. I was determined to make it work and this experience strengthened my resilience and problem-solving skills.
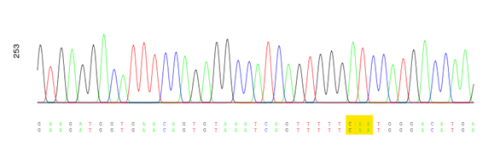
I acquired many useful laboratory skills which will be very invaluable as a researcher. I learned how to read vector maps, how to clone, use publicly available software, and how to design primers for PCR… just to name a few. Understanding the science behind molecular biology techniques allowed me to modify protocols to troubleshoot efficiently. Finally, the Sanger-sequencing results came back, and excitingly, we validate our single-point mutation H100Q of the human-NRAP gene in pBluescript II SK (+) (Figure 1).
Apart from generating the mutation, I learned how to conduct research on live animals during my work in the fish facility. I bred and cared for zebrafish embryos until up to 5 days post-fertilisation period during which they are not regulated under a Home Office licence. I practiced my microinjection skills into one-cell stage embryos. My fascination only grew stronger and I found myself peaking at the embryos throughout the day to witness the one cell splitting into 4-, 8- and 16-cells forming a plate on top of the yolk and was amazed how, in the span of only 48 hours, I could make out the heart structure and observe the heartbeat under a stereomicroscope. Another interesting aspect of this project was to investigate the H100Q-NRAP mutation in Pinniped and hypothesize its origin in Otariids and Odobenidae.
Overall, I was thrilled to discover how different aspects of biology, from molecular biology, and genetics to developmental biology and physiology came together in this project. It reminded me once again of the importance to follow a big-picture approach and that scientific understanding can seldom follow from isolated observations. The point is, however, to put them into the context of constant change interplay with each other and the environment. A continuation of this project would on the one hand be useful to further aid the scientific understanding of cold adaptations and hibernations to conserve ecosystems and species being threatened by climate change and on the other hand understand the physiological implication of heart mutations in humans.
I would like to thank my supervisor Dr. Claire Russell, Dr. Caroline Pellet-Many, Dr. Steve Allen, and Dr. Denis Larkin for their support, insights, well-stocked freezers full of enzymes, and encouragement throughout the whole process. I am also kindly thanking the British Society for Developmental Biology for granting me this fantastic opportunity and opening the doors to the world of scientific research for me. Thank you!
References
Buggiotti, L.; Yurchenko, A.A.; Yudin, N.S.; Vander Jagt, C.J.; Vorobieva, N.V.; Kusliy, M. A.; Vasiliev, S.K.; Rodionov, A.N.; Boronetskaya, O.I.; Zinovieva, N.A.; Graphodatsky, A.S.; Daetwyler, H.D; Larkin, D.M. (2021) ‘Demographic History, Adaptation, and NRAP Convergent Evolution at Amino Acid Residue 100 in the World Northernmost Cattle from Siberia’, Molecular Biology and Evolution, V(38), Issue(8), pp. 3093–3110, https://doi.org/10.1093/molbev/msab078
GeneCards (2022) NRAP gene – Nebulin related anchoring protein. Available at: NRAP Gene – GeneCards | NRAP Protein | NRAP Antibody


 (3 votes)
(3 votes)