BSDB Gurdon Summer Studentship Report (14)
Posted by BSDB, on 31 October 2017
![]() Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding.
Our third report from the 2017 group of student awardees comes from Rachael Adams (student at University of Cambridge), who undertook her studentship with Peter Lawrence at the Dept. of Zoology in Cambridge.
This summer, thanks to the Gurdon/The Company of Biologists Summer Studentship, I was fortunate to spend 8 weeks working in Dr Peter Lawrence’s lab in the Department of Zoology.
The group studies PCP, a pathway which coordinates cell polarity and helps to align epidermal patterns in the Drosophila abdomen. Drosophila larvae are covered with a cuticle decorated by denticles which help the larvae to grip substrate in order to move. These denticles form a specific pattern and changes in the pattern can be used to investigate the properties of the PCP pathway. PCP genes are highly conserved and have been identified as being involved in processes such as vertebrate gastrulation, demonstrating their fundamental importance in animal development and patterning.
My project aimed to investigate the importance of Rab GTPases to PCP. Much research has been carried out on the Rab family of proteins, as they act as master regulators of intracellular membrane trafficking. The accurate delivery of cargo between organelles is crucial for normal eukaryotic cell function. Rabs facilitate this by coordinating vesicle formation, transport and fusion.
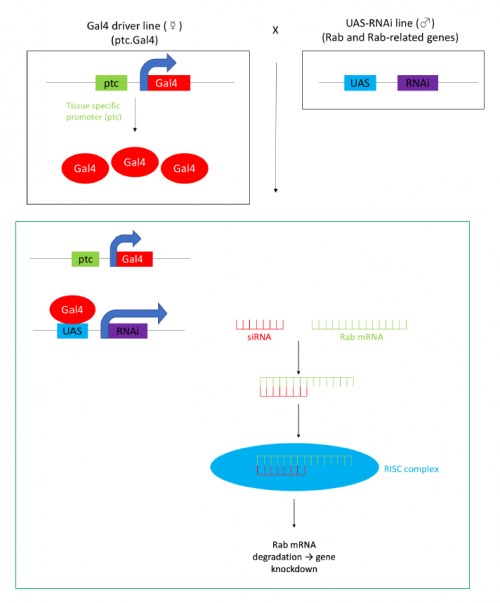
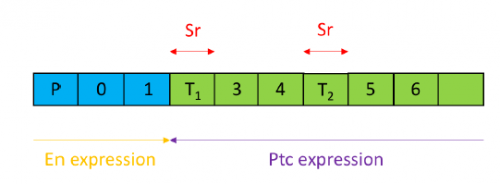
In order to test the role of as many Drosophila Rabs as possible I carried out a UAS RNAi knockdown screen of 34 different Rab and Rab-related genes, see figure 1. This screen took 7 weeks to carry out as I had to cross a collection of 103 different UAS-RNAi lines with ptc.Gal4 virgin females. This involved maintaining a population of ptc.Gal4 flies in several bottles. I crossed the virgins with males obtained from each UAS-RNAi stock and incubated the flies at 29℃, the optimal temperature to see the effect of the RNAi. Once each cross was made it was approximately 5 days before third instar larvae could be collected. To identify any change in the phenotype, I mounted several third instar larval progeny of each Gal4-UAS- RNAi cross and examined the denticle pattern produced under the microscope. Most larvae exhibited a wild type pattern, this may be because most of the Rabs I examined don’t have a function in PCP or redundancy of Rab proteins may mask certain phenotypes. The low efficacy of some of the RNAi constructs may have also affected the proportion of successful knockdowns. However, I found that several of the Rab23 crosses produced an unusual phenotype where the tendon cell gaps appeared larger than normal, see figure 2. In order to test whether this result is repeatable, I have crossed the UAS-RNAi Rab23 stocks to different driver lines (sr.Gal4 and en.Gal4). As Gal4 expression is under the control of a different promoter in each driver line, this results in a different expression pattern of the RNAi, see figure 3. Whilst different drivers will not produce the same phenotype, if the denticle pattern is disrupted it may help in the investigation of the role of Rab23 in PCP.
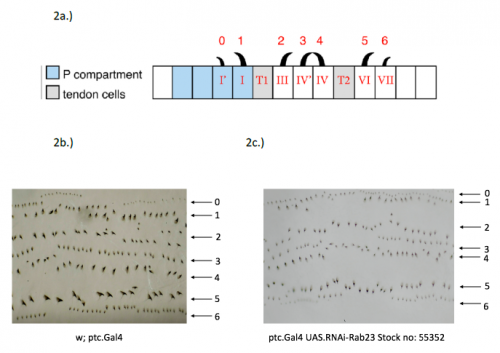
The results from the ptc.Gal4- UAS-RNAi Rab 23 crosses raised the question of why the tendon cell gap appeared larger. Possibilities include: that the tendon cells were enlarged, divided more frequently or the surrounding cells were smaller. To investigate this phenotype, I set up a series of crosses and aim to ultimately generate larvae carrying the Rab23 knockdown and expressing spaghetti squash protein (sqh) labelled with the fluorescent marker mCherry. Sqh encodes the regulatory light chain of non-muscle myosin so is expressed throughout cells. The sqh.mCherry construct therefore allows individual cells to be visualised with a confocal microscope and could help identify why the tendon cell gap is expanded.
To further investigate the phenotype I set up crosses to produce marked clones in adult flies where Rab23 is knocked down, the resultant mosaic animal allows comparison with wild type cells. This is achieved by using the Flp-FRT system to bring about site-specific recombination, see figure 4.
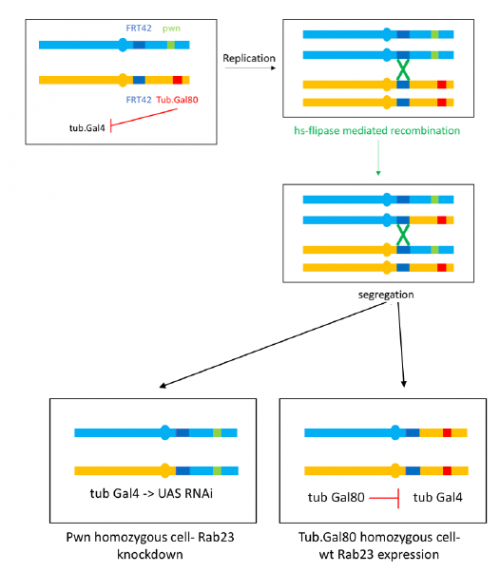
I have really enjoyed my time in the lab and have learned a great deal. I am looking forward to continuing to investigate the Rab23 phenotype and seeing the results of my crosses as part of my Part II Zoology project this term. My special thanks to José Casal for supervising and to everyone in the lab for providing a great deal of help and advice.


 (2 votes)
(2 votes)