Camelid antibodies go fishing
Posted by Paolo Panza, on 18 May 2015
Figure 1. “Cytoplasm”, illustration by David S. Goodsell, the Scripps Research Institute.
When contemplating the illustrations by David S. Goodsell (Figure 1), the first thing that stands out is how cells are packed full with those wonderful little machines we call proteins. They move, interact and change shape to produce cellular functions, so our ability to observe where they localize and monitor their behaviour is extraordinarily important in biology. In general, immunocytochemistry is preferred for static protein detection, while the dynamics of protein localization can be highlighted by transgenic constructs including the protein of interest fused to a fluorescent tag. Importantly, whereas tagged proteins are irreplaceable tools for live imaging at high resolution, immunocytochemistry is far from dead. Both antibodies and fluorescent tags are so widespread in use that sometimes we assume that the results we obtain must reflect the endogenous condition. As a matter of fact, when comparing data from static and dynamic protein detection techniques, inconsistencies can emerge. On one hand artifacts can derive from handling of the tissue, for example by fixation and permeabilization (Schnell et al., 2012). On the other hand, direct tagging of a protein or perturbations in its expression levels can interfere with the way proteins fold, interact and therefore localize. In a notable example, Swulius and Jensen (2012) found that YFP-tagged MreB (a bacterial actin homolog) artifactually nucleates to form long helical structures. The description of a helical cytoskeleton in rod-shaped bacteria heavily influenced this field of research for over 10 years. However, the aggregates were not found in microstructural analyses of bacteria with untagged MreB, demonstrating that the filaments were a byproduct of protein tagging.
In the current issue of Development, we contributed a report investigating a potential alternative for protein detection in living organisms, which unifies the advantages of antibodies and live imaging.
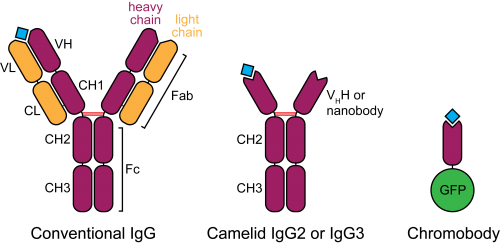
Figure 2. Schematic structure of camelid heavy-chain antibodies and chromobodies, compared to conventional IgGs.
In the late 1980s, a serendipitous discovery led to the description of heavy-chain antibodies in the plasma of camelids (Hamers-Casterman et al., 1993). Similar molecules are also found in sharks, however their advantage for animal immunity is not clear. These atypical IgGs are devoid of light chains, such that their heavy-chain variable portion (termed VHH or nanobody) can alone recapitulate antigen binding (Figure 2). This convenient property introduces the possibility of cloning entire repertoires of nanobodies and screening for binding to an antigen of interest in vitro, for example by phage display.
Nanobodies are already important reagents utilized in X-ray crystallography. They preferably bind convex surfaces, thereby helping the stabilization of protein conformations or by providing additional contact surface for protein aggregates (De Meyer et al., 2014). Their small size (15 KDa, one tenth of a conventional antibody) and high affinity are valued properties. In addition, with more and more monoclonal antibodies currently being evaluated for therapeutic application, nanobodies are becoming increasingly popular. They offer a variety of advantages, among which high tissue penetration, blood clearance, stability and solubility (Holliger and Hudson, 2005).
In a most innovative adaptation of nanobodies, Rothbauer, Zolghadr et al. (2006) fused the VHH binding moiety to a fluorescent protein and tested the resulting “chromobody” in mammalian cells. In this context, chromobodies can recapitulate the localization and original behaviour of endogenous proteins, with minimal functional interference, presumably because of their high binding turnover. Remarkably, chromobodies appear to fold correctly by establishing their only disulphide bond even in the reducing intracellular environment.
Video 1. 5 hpf zebrafish embryo injected with actin chromobody mRNA, showing localization to the plasma membrane and to the perinuclear actin cap. Centrosomes can be identified during cell division, after which the nuclear compartment and its actinic cap reassemble (arrowheads). Scale bar: 50 µm.
Inspired by these results, Ulrich Rothbauer (now at the Universität Tübingen) initiated a collaboration with our group at the Max Planck Institute for Developmental Biology, aimed at testing chromobody reagents in zebrafish. Would antigen binding be maintained in an entire organism? Would it be stable over consecutive developmental stages? We decided to focus on chromobodies binding to two proteins with well-known localization patterns, F-actin and PCNA. This choice was also motivated by the fact that interfering with actin or PCNA function would probably lead to visible phenotypes, conveniently showing side effects of chromobody expression. After injecting chromobody mRNA in zebrafish zygotes we could observe localized fluorescence at the expected sites where the respective antigens normally accumulate (Video 1). However, because of the expression variability and aspecific death in many embryos, we decided to switch to genetically-encoded expression systems. Therefore, by combining chromobodies and heat-shock or Gal4-dependent expression, we collected live imaging data supporting the actual binding of chromobodies to endogenous intracellular antigens in zebrafish, as confirmed by costaining with antibodies. With to the newly generated heat-shock and UAS transgenic lines we encountered some photobleaching problems, however we could detect actin and PCNA dynamics with high fidelity in different tissues or in the entire embryo, throughout embryonic development.
A very important question regards the impact of chromobodies in the context of a living embryo. Will they interfere with endogenous protein function? Strikingly, we observed that transgenic zebrafish embryos and larvae were morphologically normal throughout development and adult stages. They did not show aberrations in cell cycle progression (when monitoring the PCNA chromobody), nor they could alter the migration of lateral line primordium cells (in the case of the actin chromobody), even though it is known that interfering with actin dynamics at the leading edge of the primordium will perturb its migration (Xu et al., 2014). We sought a possible explanation for these observations. We detected fast recovery after photobleaching in HeLa cells stably expressing chromobodies. Importantly, the original localization of the antigen was immediately recapitulated by unbleached chromobodies, indicating that these molecules have elevated koff and kon, which imply a very transient antigen binding mode. We propose that the short binding half-life displayed by chromobodies prevents the emergence of global functional defects in living embryos.
In our work we show that chromobodies can be used as research reagents in living vertebrates, and substantially behave as already described in cultured mammalian cells. We introduce chromobodies as a complementary technique to more standard immunocytochemical and fluorescent-tag-based approaches. Chromobodies are advantageous because they allow live imaging without the need to directly modify or overexpress the protein of interest, approaches that can potentially produce artifacts. From the application side, chromobodies can be used as readout for phenotyping purposes or for high-throughput drug screening. As an example, the PCNA chromobody reports cell cycle progression just by displaying the localization dynamics of PCNA, which allows to differentiate single cell cycle stages and confers better resolution during S phase and S/G2 transition compared to FUCCI (Sakaue-Sawano et al., 2008).
Recent publications reported improved methods to clone and isolate nanobodies (Fridy et al., 2014), hence we hope that the high-throughput identification of novel binders will boost the availability of these reagents for basic research community. Although our results are preliminary because they focus only on two commercial chromobodies, we anticipate a widespread adoption of these promising reagents.
Featured article
Panza, P., Maier, J., Schmees, C., Rothbauer, U., & Sollner, C. (2015). Live imaging of endogenous protein dynamics in zebrafish using chromobodies Development, 142 (10), 1879-1884 DOI: 10.1242/dev.118943
References
De Meyer, T., Muyldermans, S. and Depicker, A. (2014). Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 32, 263–270. DOI: 10.1016/j.tibtech.2014.03.001
Fridy, P. C., Li, Y., Keegan, S., Thompson, M. K., Nudelman, I., Scheid, J. F., Oeffinger, M., Nussenzweig, M. C., Fenyö, D., Chait, B. T., et al. (2014). A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260. DOI: 10.1038/nmeth.3170
Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hammers, C., Songa, E. B., Bendahman, N. and Hammers, R. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448. DOI: 10.1038/363446a0
Holliger, P. and Hudson, P. J. (2005). Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 23, 1126–1136. DOI: 10.1038/nbt1142
Rothbauer, U., Zolghadr, K., Tillib, S., Nowak, D., Schermelleh, L., Gahl, A., Backmann, N., Conrath, K., Muyldermans, S., Cardoso, M. C., et al. (2006). Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889. DOI: 10.1038/nmeth953
Sakaue-Sawano, A., Kurokawa, H., Morimura, T., Hanyu, A., Hama, H., Osawa, H., Kashiwagi, S., Fukami, K., Miyata, T., Miyoshi, H., et al. (2008). Visualizing Spatiotemporal Dynamics of Multicellular Cell-Cycle Progression. Cell 132, 487–498. DOI: 10.1016/j.cell.2007.12.033
Schnell, U., Dijk, F., Sjollema, K. A. and Giepmans, B. N. G. (2012). Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 9, 152–158. DOI: 10.1038/nmeth.1855
Swulius, M. T. and Jensen, G. J. (2012). The Helical MreB Cytoskeleton in Escherichia coli MC1000/pLE7 Is an Artifact of the N-Terminal Yellow Fluorescent Protein Tag. J. Bacteriol. 194, 6382–6386. DOI: 10.1128/JB.00505-12
Xu, H., Ye, D., Behra, M., Burgess, S., Chen, S. and Lin, F. (2014). Gβ1 controls collective cell migration by regulating the protrusive activity of leader cells in the posterior lateral line primordium. Dev. Biol. 385, 316–327. DOI: 10.1016/j.ydbio.2013.10.027



 (3 votes)
(3 votes)
Just a footnote:
Nanobodies is a worldwide registered trademark of Ablynx (Belgium).