A Crumby affair: Cell ingression during gastrulation
Posted by Nitya Ramkumar, on 31 January 2017
Comment on “Crumbs2 promotes cell ingression during the epithelial-to-mesenchymal transition at gastrulation”
Ramkumar N, Omelchenko T, Silva-Gagliardi NF, McGlade CJ, Wijnholds J, Anderson KV.
Nat Cell Biol. 2016 Dec;18(12):1281-1291
Lewis Wolpert (1986)
Aptly stated by Wolpert, gastrulation is the fundamental process that shapes the morphogenesis of the embryo. It involves the formation of the three germ layers, namely the endoderm, mesoderm and ectoderm, from a single layer of epithelial cells called the epiblast. These germ layers then proceed to give rise to the different tissues and organs in the embryo. Gastrulation involves large scale cell movements, rearrangements and changes in gene expression leading to transformation of cell fates. Among them is a process called epithelial-to-mesenchymal transition (the EMT), wherein epithelial cells lose their apical-basal polarity and adopt migratory mesenchymal behavior. In the mouse embryo, this process happens at the posterior pole of the embryo in a structure called the primitive streak, which is also a convergence point for many signaling pathways in the embryo1. The sheer complexity of gastrulation and its efficiency of execution fascinated me and motivated me to work on it during my PhD.
The bulk of our current knowledge and understanding of the signaling pathways involved in gastrulation comes from the analysis of mutants identified from genetic screens mainly in Drosophila and recently in other animals. Likewise, my journey to the world of Crumbs and its role in gastrulation began with a mouse mutant called wsnp (wing shaped neural plate). wsnp mutants have very little mesoderm-derived tissues and their neural plate resembles the Drosophila wing disc.
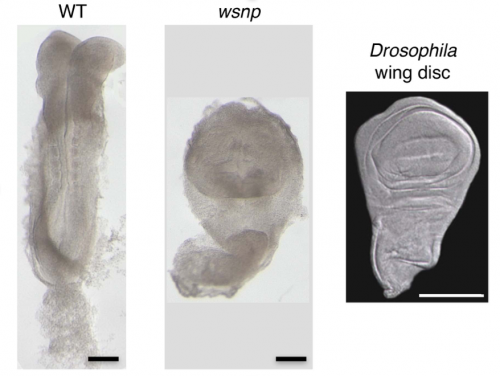
With the help of deep sequencing and genetic complementation analysis we identified wsnp to be a null allele of Poglut1 (Protein O-glucosyltransferase) which encodes an enzyme that adds O-glucose to the EGF repeats of proteins. At the time we discovered this, Drosophila Notch was shown to be modified by this enzyme2. While I worked on studying the role of Notch glycosylation during mammalian gastrulation, the severity of the wsnp phenotype compared to that of Notch pathway mutants led us to explore the possibility of alternate targets of the enzyme during gastrulation. Among them, the Crumbs family of proteins stood out as likely candidates. In the lab, we had previously isolated a mutant lulu, a null allele of Epb4.1l5, which had a phenotype similar to wsnp at gastrulation, and biochemical experiments had showed a direct interaction of Epb4.1l5 with mammalian Crumbs3,4. With this as our only inkling, we took a leap of faith and decided to explore the role of Crumbs in gastrulation. At that time, there was no precedent for the role of any Crumbs proteins in mouse development, let alone gastrulation. We decided on Crumbs2 because it had a long extracellular domain with EGF repeats that could be glycosylated and the conserved intracellular domain. We waited for about a year to get the conditional Crumbs2 knock out mice and the results made our wait worthwhile. We were really excited that the phenotypes of wsnp and Crb2-/- were identical. We went on to show that sugar modification of Crumbs2 is essential for its membrane localization and that in turn is essential for its function during gastrulation5.
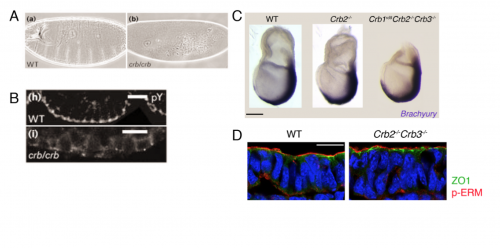
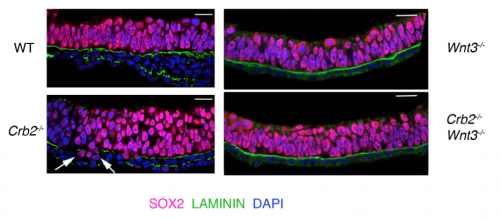
We then focused our attention to our central question – What is the function of Crumbs2 during mammalian gastrulation? We had always known Crumbs as a well-established apical determinant in Drosophila as its role in cell polarity has been well explored there6,7. We were surprised to find that the epiblast of Crumbs2-/- mutants did not have any polarity defects. To rule out the involvement of other mammalian Crumbs proteins in setting up this polarity, we generated triple mutants lacking all known mammalian Crumbs proteins. Any mouse geneticist will tell you – generating triple mutants is not fun! To our surprise, we found that mammalian Crumbs proteins were not required to establish the polarity of the epiblast epithelium. This was in contrast to the case in Drosophila, where Crumbs is essential to set up polarity of the embryonic epithelium. Additionally, in the Crumbs2-/- mouse embryos we found an accumulation of cells expressing E-cadherin and Sox2+at the streak, suggesting that these cells were still epithelial. This was contrary to what we were expecting: as a canonical polarity protein, one would imagine that a loss of Crumbs2 would lead to more cells delaminating i.e. making more mesoderm cells, but we saw the opposite. This suggested to us that the functions of mammalian Crumbs2 could be different from that of Drosophila Crumbs, and got me further engrossed into understanding how mammalian Crumbs2 functions during gastrulation.
Our attention was drawn to the cells stuck at the streak. Although we were able to visualize these cells using SEM and X-linked GFP, these were static, 2-dimensional images. Cell delamination from an epithelium is a dynamic process requiring constant neighbor exchange and happens in 3-dimensional epithelium. In order to understand exactly when the cells fail during this process, we had to watch this process live in mouse embryos. While it seemed like the most obvious experiment, it was clearly the most challenging aspect of the project at many levels. At that time, there were hardly any labs that had done live imaging with fluorescent reporters in mouse embryos. While cell delamination had been extensively studied, and imaged in in vitro culture systems, only recently with the advent of better imaging technology and improved reporters scientists have started exploring it in live tissues and embryos. For us, it was a herculean task as we had to work out every aspect of the experiment including the mouse strains to use, then determine the imaging conditions to use while keeping the embryo stable. Additionally, we had to define the temporal dynamics of this process in wild-type embryos before we begin to compare it with our mutant.
As mouse embryos are very sensitive to laser light and the primitive streak is a densely packed epithelium, it is important for the reporter to be very bright to get reasonable data. Initially we tried with reporters expressed ubiquitously in the epithelium, as it would reduce the number of crosses and increase the probability of the desired genotype. However, at that time there weren’t good algorithms to segment these images making their analysis very difficult and unfruitful. To bypass this, we decided to express the fluorescent protein in a random pattern, which helps to create better 3-dimensional reconstructions of individual cells and help in their subsequent analysis. However, this would involve additional crosses and also decrease the probability of getting our desired genotype. After screening many different reporter and CRE lines that we and others at Sloan Kettering had in their labs, we began to lose hope in this experiment, as we weren’t getting the right combination of the two. It was around this time that Kathryn attended a conference where Ann Sutherland presented her lab’s work on imaging with the mT-mG reporter and the EIIA-CRE. They found that this combination gave them a good random pattern and the membrane-GFP (mGFP) was a bright reporter at this stage8. Having seen her beautiful movies with the mGFP, we decided to get these mice and eventually used them for our experiment. Despite our optimization of the imaging, there were many factors in this experiment that were beyond our control. For instance, the probability of getting a mutant with both the reporter and the CRE was low and the percentage of randomness of the CRE was unpredictable. Nevertheless, after many unsuccessful attempts, we managed to get beautiful movies of the cells in wild-type and the mutant streak and were happy to see that the cells in the mutants shrink their apical surface but fail to leave the epithelium. While we focused mainly on shape changes and time to delamination for the purpose of this paper, these movies are a gold mine of data. It is amazing how so many processes are happening simultaneously at the streak. There are cell divisions, tissue level movements, neighbor exchanges to just name a few. With the advent of light sheet microscope, there is undoubtedly lots to learn from just watching these cells live in the epithelium.
GFP positive streak cell translocates from the apical to the basal side of the epiblast in wild type embryo. The epiblast cells in the mouse embryo randomly express mGFP at E7.5 and are imaged from the primitive streak side (posterior). A 3D rendered surface (yellow) was built with Imaris for easy visualization of the individual cell shape changes. Ingressing cells have basal protrusions, translocate their cell body basally and leave the epiblast in less than 2 h. In addition to the shape changes we discuss in the delaminating cell, the other cells in this region of the epithelium (streak), which are poised to delaminate are also highly dynamic. Time is h:min. Scale bar,10 µm.
The big debate- Polarity?
Since Crumbs was as a polarity protein, we had to determine whether the defect we saw was specific to gastrulation at the streak or that Crumbs plays a role in the epithelia by virtue of which it affects the streak. My lab meetings would always end on this debate, followed by heated arguments supporting both hypothesis. The only way to test this was how Drosophila geneticists had always done before. We had to prevent gastrulation from happening using Wnt3 mutants (with and without Crumbs2) and see whether the tissue was normal or not. Fortunately for us, we had the right tools out there to do this experiment. Since both outcomes were equally likely and exciting, irrespective of the result, it was one of the most exciting dissections ever. It would answer the question I had since my first lab meeting. I remember discussing the Wnt3 results at my lab meeting: finally, we concluded without any argument that it is the streak, and not the epiblast epithelium, that requires Crumbs2, after all.
Perspective makes a difference
Traditionally, protein localization in the primitive streak epithelium was observed by immunostaining transverse sections of the epithelium. We too followed suit to look at Crumbs2 expression and were happy to see it localized apically in the cells of the streak. During my trials with live imaging, I began to appreciate the importance of the 3-dimensional nature of the epithelium and thought it was essential to see where Crumbs2 would fit in this 3-D context. The apical surface of the primitive streak epithelium faces the interior of the embryo, making it difficult to access and therefore not many people have looked at polarity proteins en face, in contrast to studies in Drosophila and cell culture. While learning to do en face imaging on neural epithelium, as a fun experiment I decided to look at Crumbs2 localization en face at the primitive streak. Although it took me many trials to get the streak epithelium flat, I was surprised to see that Crumbs2 localization was anisotropic. At first, I thought it could be an artefact of my sample processing. However, after repeated attempts with different approaches to sample processing and appropriate controls I was finally able to convince myself about the anisotropic pattern. Nevertheless, we still didn’t understand what it meant! I had tried co-staining with many other polarity proteins, but none of them had this pattern. I had immunostained for MyosinIIB separately, but it wasn’t until the Roper paper on Drosophila Crumbs in the salivary gland that we decided to probe for MyosinIIB and Crumbs2 together. That’s when it came together. The complementary nature of their pattern was striking and steered us to think of possible mechanisms of Crumbs2 action.
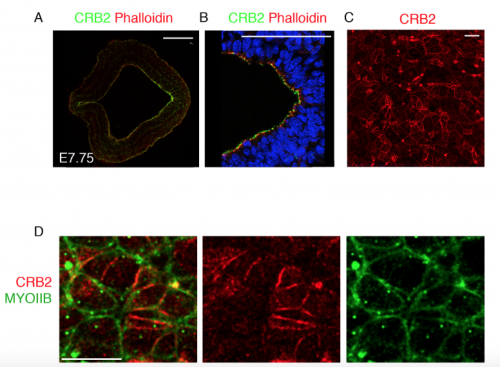
Transverse section of the primitive streak of wild-type embryo at E7.75 immunostained for CRB2 (green) and Phalloidin (red) showing the expression of CRB2 in both the anterior presumptive neural epithelium and the epiblast and its enrichment in the posterior epiblast. The streak region is magnified in (B). Extended projection of en face view of the apical surface of the epiblast layer of the primitive streak of wild type at E8.5 immunostained for CRB2 (C), showing its anisotropic distribution. The double immunostaining of the streak for CRB2 (red) and MYOSIN IIB (green), showing the reciprocal enrichment of CRB2 and Myosin IIB on different cell edges. Proximal is up. Scale bars: A, B, 100 μm; C,D 10 μm.
We hypothesize that the unequal levels of Crumbs2 in the epithelium must be an important part of the mechanism to determine which cells delaminate first. It’s a very interesting problem which hasn’t received its due. The streak region as defined by the basement membrane breakdown is easily around 4-5 cells wide and runs from the node to the distal tip of the embryo. Despite that, cells delaminate individually and leave the epithelium, suggesting that this process must be highly coordinated and tightly regulated at both cell and tissue levels. We know little about how the cells establish the complementary pattern of Crumbs2 and MyosinIIB, how they define the order in which they delaminate and how they coordinate this with their neighbors. These questions can be addressed by live imaging with appropriate reporters and will offer a wealth of information.
My journey to Crumbs emphasizes the power of genetic screens in identifying new players in gastrulation, and my paper offers a peak into the unexplored territories of mammalian gastrulation. There is a lot to learn by just observing this beautiful process in wild-type and mutant embryos. Hopefully, my work will inspire people to explore the dynamics of this process in greater detail.
References
1 Ramkumar, N. & Anderson, K. V. SnapShot: mouse primitive streak. Cell 146, 488-488 e482, doi:10.1016/j.cell.2011.07.028 (2011).
2 Acar, M. et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132, 247-258, doi:10.1016/j.cell.2007.12.016 (2008).
3 Lee, J. D., Silva-Gagliardi, N. F., Tepass, U., McGlade, C. J. & Anderson, K. V. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development 134, 2007-2016, doi:10.1242/dev.000885 (2007).
4 Gosens, I. et al. FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp Cell Res 313, 3959-3970, doi:10.1016/j.yexcr.2007.08.025 (2007).
5 Ramkumar, N. et al. Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2. PLoS Genet 11, e1005551, doi:10.1371/journal.pgen.1005551 (2015).
6 Klebes, A. & Knust, E. A conserved motif in Crumbs is required for E-cadherin localisation and zonula adherens formation in Drosophila. Curr Biol 10, 76-85 (2000).
7 Tepass, U. Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila. Dev Biol 177, 217-225, doi:10.1006/dbio.1996.0157 (1996).
8 Williams, M., Burdsal, C., Periasamy, A., Lewandoski, M. & Sutherland, A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev Dyn 241, 270-283, doi:10.1002/dvdy.23711 (2012).


 (6 votes)
(6 votes)