A Day in the Life of a Coral Lab
Posted by YuuriY, on 8 November 2016
Hi, I’m Yuuri Yasuoka, a postdoc in the Marine Genomics Unit at the Okinawa Institute of Science and Technology Graduate University (OIST). Okinawa is a subtropical Japanese island surrounded by beautiful coral reefs (Figure 1). Why do we study coral here? OIST is the best place in Japan to study coral, with the good access to wild coral reefs and advanced research facilities for molecular biology.
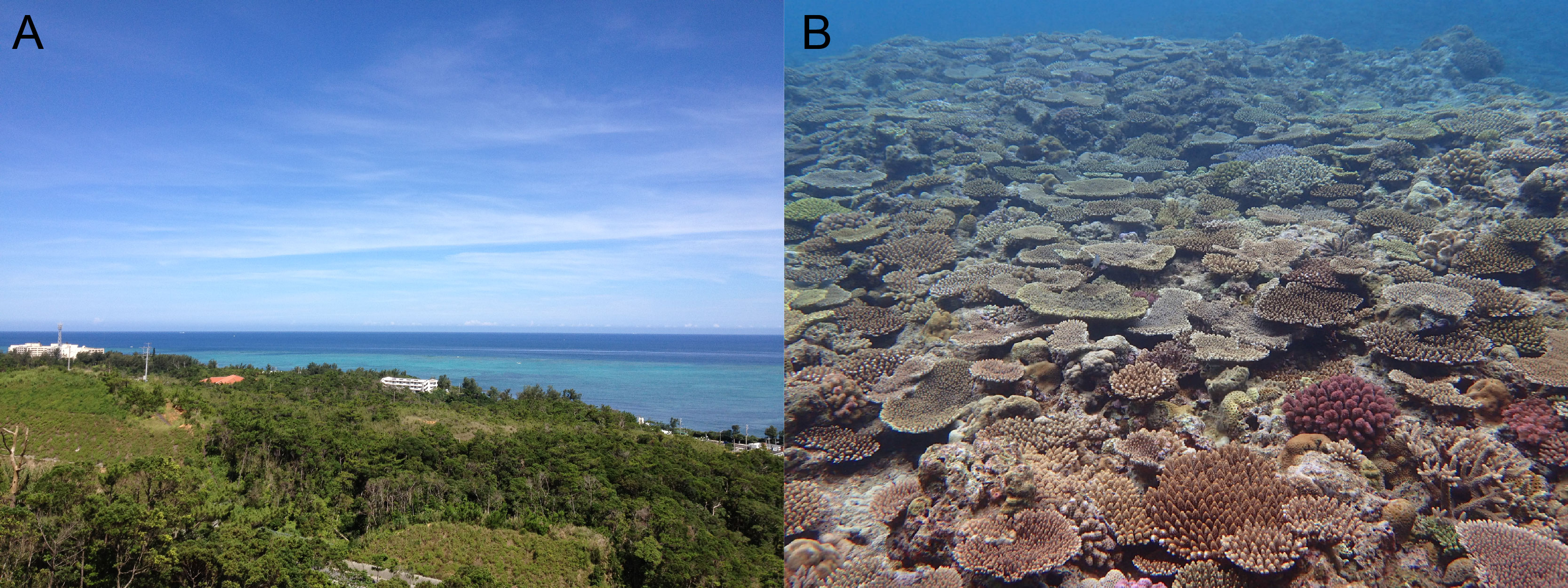
Figure 1. Beautiful coral reefs in Okinawa. (A) An ocean view from OIST. (B) Acropora community at Ishigaki Island, Okinawa Prefecture, Japan (Photo: Yuna Zayasu).
How important is it to study corals?
Coral reefs are one of the most biodiverse ecosystems in the world, and are believed to support roughly 25% of all marine species (Knowlton et al., 2010). In addition, they are also important for the local Okinawan economy, as fisheries and tourist attractions. Therefore, our unit, led by Prof. Noriyuki Satoh, has been studying coral genetics to understand coral ecosystems at the molecular level. In 2011, our group reported the first draft genome of a stony coral, Acropora digitifera (Shinzato et al., 2011). This species is very common in Okinawa (Figure 2A). Using the genome database, we can now address many biological questions concerning corals that were impossible to answer previously.
Corals occupy an important phylogenetic position when it comes to understanding evolution of metazoan body plans. Corals belong to the Phylum Cnidaria, which also includes sea anemones, hydras, and jellyfishes. It forms a sister group to the Bilateria. Although most cnidarians are marine organisms, the most widely used experimental model cnidarians, Hydra and Nematostella, are very tiny freshwater and brackish water animals. While corals are a primitive clade of cnidarians that diverged from sea anemones ~500 Mya, they are one of the most successful cnidarian groups (Shinzato et al., 2011). Thus, coral studies tell us much about conserved ancestral features of cnidarian body plans. Recently, we published a research paper analyzing molecular functions of the brachyury gene in coral embryos (Yasuoka et al., 2016). Because cnidarians lack mesoderm and because brachyury functions as a mesoderm-forming gene in vertebrates, this study sheds light on evolutionary origins of vertebrate mesoderm.
Next, I will discuss the most exciting day in a coral lab. That occurs when corals spawn.
Coral spawning
It is generally believed that corals spawn once per year under a full moon; however, in reality it is not that simple in Okinawa. First, spawning is totally unpredictable. It ranges from one week before a full moon to two weeks after. For spawning experiments, we collect 5-10 wild coral colonies about two weeks before experiments and keep them in seaside aquaria. Every night, we check them to see whether they show signs of spawning. This is called “bundle setting,” as corals produce red or orange bundles of gametes near the mouths of their polyps (Figure 2B). In Acropora digitifera, this occurs at approximately 8 pm, and the colony starts to spawn at around 10 pm (Figure 2C). Interestingly, another Acropora species, A. tenuis starts to spawn at around 8 pm. The spawning time differs between species, but it is very precise.

Figure 2. Coral spawning. (A) Acropora digitifera colonies in shallow water at Onna-son, Okinawa, Japan. (B) “Bundle setting” of Acropora tenuis (Photo: Yuna Zayasu). (C) Acropora digitifera colonies that are spawning.
Second, the spawning day is highly variable, depending on location and local conditions. Acropora species spawn during May and June. Usually, corals in the Yaeyama Islands, which are located in southwestern Okinawa Prefecture, spawn one month earlier than at Okinawa Island. Even around the same island, the spawning day differs between locations. In fact, among our colonies in aquaria, collected at the same location, not all colonies spawned on the same night. Furthermore, we have observed that some colonies spawn twice in one season. Thus, there are many challenges in obtaining coral gametes, but with considerable experience, we have become quite successful.
Movie 1. Spawning of Acropora digitifera.
Microinjection of coral eggs
If we were not coral researchers, we would be satisfied just to be able to observe the beautiful and mysterious spawning of corals (Movie1). However, after spawning occurs we have to do our experiments. Acropora corals spawn bundles that contain both eggs and sperm. After bundles float to the water surface, the surrounding membrane ruptures to release eggs and sperm. Because they do not self-fertilize, we collect bundles from different individuals separately to perform in vitro fertilization (Figure 3A). After collecting bundles from seaside aquaria, we bring them to the lab at OIST by car; it takes ~15 min. When they arrive at the lab, the collecting tubes include an orange egg layer on the surface and white sperm solution (Figure 3B,C). To fertilize the eggs, we mix eggs and sperm from different individuals. In our experience, Acropora gametes can actively fertilize at least as much as 5 hours after spawning. Thus, we can obtain synchronously developing embryos at 30 min ~ 1 hour intervals.

Figure 3. Gamete bundles of Acropora. (A) Collection of gamete bundles from each spawning individual separated in buckets at Sesoko Marine Station of the University of Ryukus, 2012. (B) Eggs and sperm separated in a collecting tube. (C) Magnified image of bundles containing both eggs and sperm.
We start microinjections 30 min after fertilization. Because Acropora eggs are very yolky and float on sea water, we had to develop a method to immobilize them (Figure 4). First, fertilized Acropora eggs are positioned between glass capillaries attached to a glass slide. Next, sea water is removed as much as possible, resulting in deformation of embryos due to the surface tension of the remaining water. Then, we inject antisense morpholinos into immobilized embryos to inhibit functions of specific genes. After microinjection, embryos are returned to sea water where they develop normally. Usually, I inject 100-200 embryos from a single batch for each type of morpholino; this takes 30-45 min. When I finish an experiment for several kinds of morpholino, it is usually 3:00-4:00 am. After microinjection experiments, I drive home with tired eyes. Injected embryos are sorted based upon fluorescence of injected materials and used for further experiments.
After numerous trials and errors, I developed this method and completed the first analysis of gene functions during coral development (Yasuoka et al., 2016). Because we can do experiments using coral embryos only once per year, it took five years to publish the paper.
Figure 4. Microinjection experiments of coral eggs. (A) Equipment for coral egg microinjection. (B) Schematic of our microinjection method. (C) Acropora embryos aligned between glass capillaries. (D) A photo seen with a stereomicroscope, showing embryos are deformed and retained by the surface tension.
Future perspective of coral studies
One of the biggest concerns about coral reef ecosystems is “coral-bleaching.” Inside their cells, corals possess symbiotic algae (Symbiodinium), on which they depend for photosynthetic products. If corals suffer severe stresses, such as high temperature, acidification, or oxidation, the symbiotic relationship collapses and corals bleach and die. Coral bleaching is an increasingly serious problem around the world, due to global climate change. However, the genetic basis of this symbiosis is largely unknown. In 2013, our lab also decoded the genome of a species of Symbiodinium (Shoguchi et al., 2013). Using the microinjection technique, now we are trying to determine the molecular mechanisms underlying coral-bleaching.
References
Knowlton, N., Brainard, R. E., Fisher, R., Moews, M., Plaisance, L., & Caley, M. J. (2010). Coral reef biodiversity. Life in the World’s Oceans: Diversity Distribution and Abundance, 65-74.
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., Fujie, M., Fujiwara, M., Koyanagi, R., Ikuta, T., Fujiyama, A., Miller, D. J., & Satoh, N. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature, 476(7360), 320-323.
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., Takeuchi, T., Hisata, K., Tanaka, M., Fujiwara, M., Hamada, M., Seidi, A., Fujie, M., Usami, T., Goto, H., Yamasaki, S., Arakaki, N., Suzuki, Y., Sugano, S., Toyoda, A., Kuroki, Y., Fujiyama, A., Medina, M., Coffroth, M. A., Bhattacharya, D., & Satoh, N. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Current Biology, 23(15), 1399-1408.
Yasuoka, Y., Shinzato, C., & Satoh, N. (2016). The mesoderm-forming gene, Brachyury, regulates ectoderm-endoderm demarcation in the coral, Acropora digitifera. Current Biology, 26(21), 2885-2892.



 (7 votes)
(7 votes)
This is a very cool and informative post.
Super cool and informative post. I am already looking for the material to build my own sponge-embryo-injecting-chamber! Thanks for sharing and congrats for your awesome work!