Fat to the forefront of histone regulation
Posted by Matthew R Johnson, on 21 August 2018
All life requires energy. For early metazoan development, demand is especially high, as the transition from a single cell to a complex, multicellular organism requires a massive energetic input. In the earliest stages of development, however, an organisms’ inability to feed poses an apparent problem: how is the energy necessary to drive development obtained? In most species, this problem is solved via maternal contribution. In Drosophila, for example, newly laid embryos contain abundant neutral lipids that are broken down to generate much of the energy necessary to drive embryogenesis: prior to egg laying, these neutral lipids are deposited into the egg by the mother, in the form of lipid droplets (LDs). While the critical roles of LDs in energy homeostasis have been investigated for decades, recent research has demonstrated that LDs may serve moonlighting functions beyond lipid metabolism, including roles in protein handling.
The lipid droplet proteome is vast, containing a variety of proteins. Many of these proteins, as one might expect, have central roles in lipid metabolism. However, what may come as a surprise, is the observation that several LD-resident proteins have known functions outside of lipid homeostasis. For years, such proteins were thought to be mere contaminants of a “dirty” LD purification procedure. More recently, however, a growing field of research has demonstrated that such localization is not only real, but also can have profound impacts on protein abundance and function.
Our story began just over a decade ago when our group and their collaborators identified, with high confidence, hundreds of proteins localized to the surface of LDs in early Drosophila embryos1. Of particular intrigue was the identification of a subset of histone proteins, namely core histones H2A and H2B as well as the Drosophila H2A variant H2Av. A priori, the obvious explanation was that these abundant, highly charged histones were likely the result of contamination, yet the absence of other histones e.g., H3 and H4, suggested a radical alternative: H2A, H2B, and H2Av are specifically recruited to the surface of LDs in Drosophila embryos, a hypothesis we later confirmed via immunohistochemistry and live imaging. This surprising finding begged the question “how are histones specifically recruited to the LD surface and, most importantly, why?”
A follow up study by our group identified the novel protein Jabba as the anchor necessary to recruit H2A, H2B, and H2Av to LDs2. To investigate potential functions of such recruitment, a previous member of the lab knocked out Jabba and discovered that histones were not only gone from LDs, but that global histone levels were also dramatically reduced. The group hypothesized that histone binding to LDs therefore allows high levels of histone to be stably maintained in the embryo, protected from surveillance mechanisms/degradation, yet made available for packaging during the rapid rounds of DNA replication occurring in early embryos. Indeed, when new histone biosynthesis is impaired, Jabba (i.e. histones on LDs) is necessary for proper development, and in its absence, embryos show phenotypes reminiscent of DNA damage.
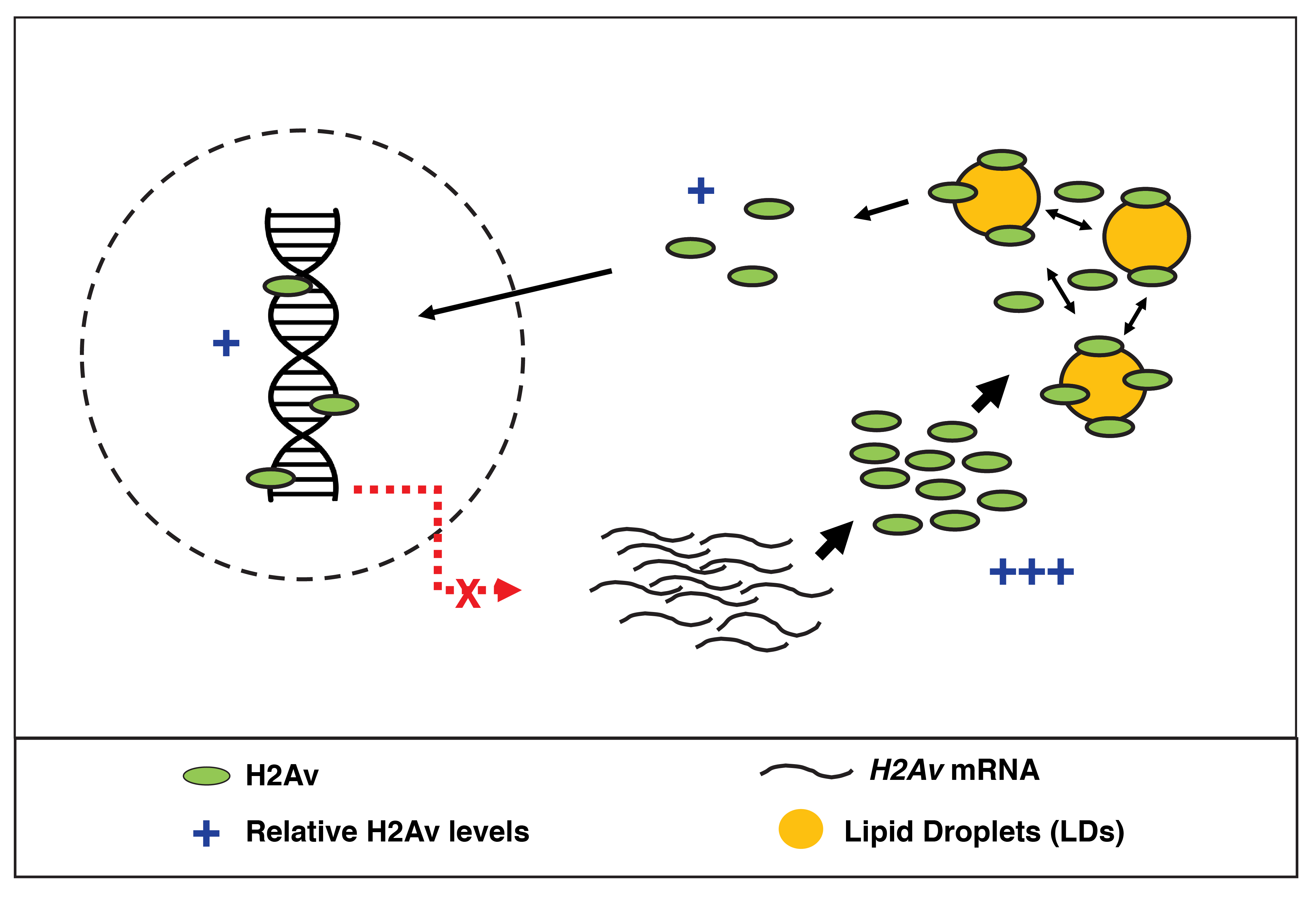
During the investigation of Jabba mutants, our group identified an intriguing phenotype suggesting that Jabba/LDs may serve an additional function in histone regulation: in Jabba embryos, the histone variant H2Av over-accumulates in nuclei, while nuclear H2A and H2B are not obviously altered3. This finding provided a paradox: on the one hand, LDs store histones to be used during development; on the other hand, they also prevent histones from entering nuclei. Could LDs serve two distinct roles in histone regulation, storing some histones for use (i.e. H2Av and H2B) while restricting others from being imported into nuclei (i.e. H2Av)? Follow-up studies showed that newly synthesized H2Av can indeed be recruited to LDs in vivo, leading us to propose that LDs act as H2Av buffers, sequestering H2Av synthesized in excess to prevent over-accumulation in nuclei. The mechanism by which buffering is achieved, however, remained unknown.
Our latest study, published last month in eLife, began when we set out to test a fundamental aspect of our histone storage model: if histones are stored on LDs for use during early development, they must then be able to leave LDs and translocate to the nucleus. Using photoactivation, a technique in which we can turn on fluorescent signal within a distinct region to allow for tracking of a particular protein population, we showed that H2Av on LDs can indeed transfer to nearby nuclei in an undisturbed, living embryo. We were particularly struck by one of the results of this experiment, namely by how fast H2Av was lost from LDs: within just minutes, nearly all of the fluorescent H2Av dissipated from the region of initial activation. We hypothesized that such rapid loss from LDs must reflect embryonic demand: in late syncytial blastoderm stages, thousands of nuclei simultaneously undergo replication, creating an immense need for histones that can be supplemented by the supply stored on LDs. To test this notion directly, we quantitated H2Av loss from LDs in earlier stages, when nuclear number and, presumably, histone demand is greatly reduced. To our astonishment, we found H2Av to be lost from LDs with similar dynamics, starkly contrasting our model that loss from LDs reflects demand.
If H2Av is always rapidly lost, despite dramatic changes in nuclear number, where is all of the H2Av going once lost from LDs? To answer this question, we took advantage of a photo-switchable H2Av-Dendra2 that we had generated: this genetic tool would allow us to unambiguously track distinct H2Av populations within the embryo. We discovered that H2Av lost from LDs was not exclusively destined for nuclei after all, but rather re-localized to neighboring LDs. At first, this constant “shuffling” of H2Av between LDs was puzzling, but we then considered whether such behavior may underlie the mechanism of our previously proposed buffering model: H2Av is re-routed back and forth between LDs, thus limiting the free pool in the cytoplasm at any given moment while simultaneously keeping H2Av available for eventual transport to the nucleus.
Based on a number of experimental observations, we developed a formal kinetic model for buffering, incorporating principles of thermodynamics and a few simplifying assumptions; this allowed us to make several key predictions, which we then sought to test experimentally. Using a variety of genetic and microscopic approaches, we were able to demonstrate that LDs are the main regulator of both cytoplasmic and nuclear H2Av. First, we showed that H2Av levels are under limited regulation in the early embryo: manipulating H2Av gene dosage was sufficient to cause a corresponding change in both global and nuclear H2Av levels. This finding was surprising, as the levels of canonical histones are kept constant by elaborate feedback mechanisms despite huge variations in gene copy number4. Second, we altered LD buffering capacity by changing Jabba gene dosage. As predicted, as Jabba levels are increased, nuclear H2Av accumulation is reduced. Third, we used FRAP to show that increased Jabba dosage is sufficient to reduce nuclear import rates of H2Av, in a similar fashion as predicted by our quantitative model.
Limiting H2Av availability via dynamic sequestration to LDs is an elegant way to prevent over-accumulation in the nucleus on short time scales. However, we reasoned that such a mechanism may prove problematic during later stages, when an elongated cell cycle provides ample time for H2Av to enter nuclei (i.e., total nuclear import of H2Av may be too high during long cell cycles, even with buffering). For example, at the time of the mid-blastula transition (MBT), interphase length is dramatically increased (~3x as long as the preceding cell cycle). We asked whether H2Av dynamics were altered during this time and discovered that H2Av becomes statically sequestered to LDs; a transition that occurs within just minutes and is dependent upon the nuclear:cytoplasmic ratio, a well-established molecular “clock” that regulates the timing of MBT events. We speculate that once the rapid cell cycles of the early embryo are finished, it becomes necessary to restrict buffering and allow canonical regulatory mechanisms to take over; investigation into the mechanism of this transition are ongoing and will allow for a direct test of this hypothesis.
To our knowledge, our work over the past several years uncovering the role of LDs in histone regulation provides the most thoroughly characterized example yet supporting a role for LDs in the handling of proteins from various cellular compartments. Furthermore, our work strongly suggests that LDs represent a novel class of histone chaperone and, in the case of the Drosophila embryo, serve as the major regulator of global and nuclear H2Av levels: such a simplified system for histone regulation is remarkable, as most cells employ a variety of mechanisms to control histone levels, including transcriptionally, post-transcriptionally, and post-translationally. In early embryos, the rapid rounds of DNA replication/cell division (~8-15 min per cell cycle) may not allow for canonical regulatory mechanisms to be effective, thus necessitating a simplified means of regulation post-translationally (i.e. buffering): whether buffering exists at other developmental stages remains an open question.
Although we do not yet know if organisms other than Drosophila employ LDs in a similar manner as regulators of histones, histone localization to LDs has been demonstrated in several other organisms/cell types, including mouse oocytes and human cancer cell lines5,6. Now that we have characterized the mechanism by which histones are regulated by LDs, it should be examined whether similar regulation exists for other LD-associated proteins; as LDs are ubiquitous organelles and LD proteomes from many organisms reveal “unusual” proteins, such a regulatory strategy may be widespread. What is clear, however, is that the functions of LDs and their implications for development and cell biology may be far more complex than once thought.
Johnson MR, Stephenson RA, Ghaemmaghami S, Welte MA.
DOI: 10.7554/eLife.36021
References:
- Cermelli S, Guo Y, Gross SP, Welte MA. 2006. The lipid-droplet proteome reveals that droplets are a protein storage depot. Current Biology 16:1783–1795.DOI: https://doi.org/10.1016/j.cub.2006.07.062
- Li Z, Thiel K, Thul PJ, Beller M, Ku¨ hnlein RP, Welte MA. 2012. Lipid droplets control the maternal histone supply of Drosophila embryos. Current Biology 22:2104–2113. DOI: https://doi.org/10.1016/j.cub.2012.09.018
- Li Z, Johnson MR, Ke Z, Chen L, Welte MA. 2014. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Current Biology 24:1485–1491. DOI: https://doi.org/10.1016/j.cub.2014.05.022
- McKay DJ, Klusza S, Penke TJ, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD, Strahl BD, Duronio RJ, Matera AG. 2015. Interrogating the function of metazoan histones using engineered gene clusters. Developmental Cell 32:373–386. DOI: https://doi.org/10.1016/j.devcel.2014.12.025
- Kan R, Jin M, Subramanian V, Causey CP, Thompson PR, Coonrod SA. 2012. Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Developmental Biology 12:19. DOI: https://doi. org/10.1186/1471-213X-12-19
- Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, Nomura DK, Olzmann JA. 2018. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Developmental Cell 44:97–112.DOI: https://doi.org/10.1016/j.devcel.2017.11.020


 (2 votes)
(2 votes)