Featured Resource: Daniocell
Posted by Jeffrey Farrell, on 20 February 2024
What is Daniocell?
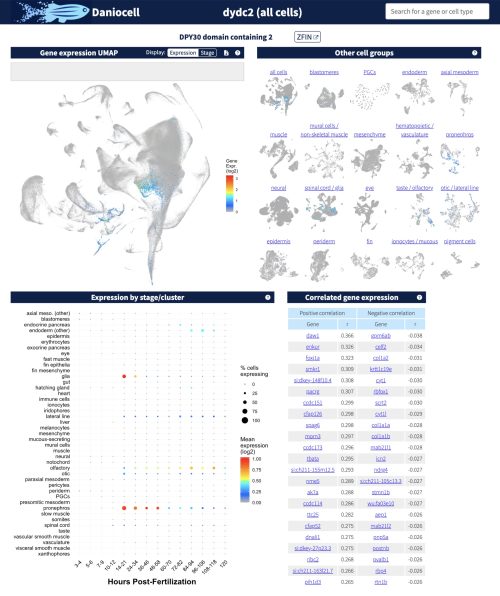
Daniocell is an exploratory tool that allows users to navigate pre-computed, time-resolved gene expression information for each cell type during zebrafish development. We profiled embryos across the first 5 days of development, as the animal progresses from a field of unpatterned cells to a swimming, feeding, and behaving animal. Daniocell showcases gene expression information for each gene in the zebrafish genome across most zebrafish cell types as they develop. For users interested in specific cell types at different stages of their development, Daniocell also holds cell type-specific pages that contains information on the most specific and most highly expressed genes within each cell type.
What inspired the development of Daniocell?
Single-cell genomics studies generate massive amounts of information, much of which is beyond the authors’ analytical goals, but could be highly useful to other members of the community. For instance, in our study, we focused on two tissues and made discoveries about several under-characterized cell types, but there are 17 other tissues that remain! Not every scientist wants to become an expert in analyzing these types of data. So, we made a list of the questions that we thought were most frequently addressed from these types of data and built a website to answer them. We hope that sharing this data with everyone will accelerate discovery-driven research within the community.
Daniocell was created from the perspective of active developmental biologists; we were hoping to create the resource we wished to have access to while performing our studies. We also tried to address feedback that we received about portals from our previous studies to try to make this one even more useful. We made some trade-offs in its development: the results are all pre-computed, which means that users cannot generate arbitrary plots to address specific questions; on the other hand, it means the website is fast, responsive, and inexpensive to run, hopefully allowing us to fund its support long-term.
How can scientists use Daniocell in their research?
Daniocell was designed to quickly and easily answer the most common questions we expect researchers might want to ask from our data, namely: (1) When and where is any given gene expressed? (2) Which genes have the most similar and dissimilar gene expression patterns? (3) When is each cell type present and undergoing the cell cycle? and (4) Which genes does each cell type express most strongly and most specifically, and how does that compare to related cell types?
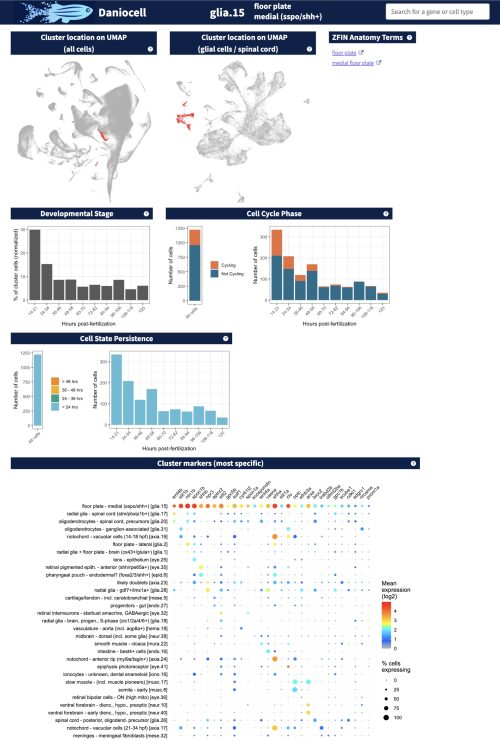
To this end, each gene page in Daniocell demonstrates (1) that gene’s expression pattern (as UMAP and dot plot) across each tissue and cell type, resolved by time, and (2) which genes have the most similar and dissimilar expression patterns across all cells and each tissue. Additionally, each cell cluster page demonstrates (1) the developmental stages of cells in the cluster, (2) whether they are undergoing the cell cycle (based on their expression of cell cycle-associated transcripts), and (3) the most specific and most highly expressed genes within each cluster.
Of course, researchers might want to generate plots with curated sets of genes or groups of cells. To facilitate this simple/easy reuse of the data, we have the fully processed Seurat object posted on the Daniocell front page, along with our annotations, so that researchers can jump right in. These data can also be used with label transfer approaches, to transfer our annotations onto newly generated scRNAseq data; this process isn’t perfect, but can accelerate progress through analyzing scRNAseq data generated across similar stages from different perturbation conditions.
Who are the people behind the resource?
This resource was designed by the Farrell Lab at NICHD/NIH. The primary people behind Daniocell were Dr. Jeffrey Farrell (PI) and Dr. Abhinav Sur (postdoc).

Abhinav Sur: I began working on zebrafish in 2020 when I started my postdoc in the Farrell Lab at NICHD. My Ph.D. was on Evodevo where I studied the evolution of nervous systems using an annelid called Capitella teleta. I generated one of the first single-cell atlases in an annelid and first single-cell atlas of Capitella teleta. The most exciting part of working on Daniocell was that it was like a “treasure hunt”, as I dug through this large dataset to uncover hidden biology. The fact that the community have found Daniocell to be a useful resource has made this experience quite rewarding. In future, I hope to continue using single-cell genomics approaches to understand how cell types are constantly produced at the right time and place in constitutively regenerating tissues such as the intestine and how their dysregulation causes disease.
Jeffrey Farrell: I’m the head of the Unit on Cell Specification and Differentiation in the NICHD Intramural Research Program. I’ve been studying developmental biology for 17 years and started using zebrafish as a research model organism a little over a decade ago. Daniocell is a continuation of my long-term efforts to profile zebrafish development using single-cell genomics approaches and to thereby uncover interesting biology that was previously missed to study using both modern and classical approaches. For me, one of the most fun parts of this project has been to hear from many of you how Daniocell has helped you in your own research.
How can researchers help and contribute to the resource?
We are very happy to receive suggestions about improvements to the annotations and metadata from members of the community, based on either newly generated information or other labs’ expertise in particular tissues. These should further increase the value of the gene expression information in Daniocell over time. They can be submitted via email, and there is a template for the information to include on the Daniocell front page.
What are the next steps for Daniocell?
We plan to update Daniocell 1–2 times per year based on community input into cell annotations and to keep it up to date with new releases of the zebrafish genome. Additionally, we hope to add a tutorial to the site demonstrating how to use the Daniocell dataset with label transfer approaches to facilitate draft annotations of newly generated scRNAseq data. We are also exploring the possibility of integrating some additional datasets into the portal or adding a more interactive component that could be used to generate custom visualization figures from the data.


 (3 votes)
(3 votes)