From local communication to global pattern recovery: spatio-temporal scales in segmentation clock and scientists
Posted by Koichiro Uriu, on 30 June 2021
By Koichiro Uriu, Bo-Kai Liao, Andrew C. Oates and Luis G. Morelli
How local cell-cell communication can generate a global tissue pattern is one of the fundamental questions in developmental biology. Yet, studying this remains challenging, because developing tissues involve complexities such as cell rearrangement, heterogeneity along the body axis, and massive tissue shape changes. In our recent paper in eLife (Uriu, Liao et al. 2021), we addressed this question using the zebrafish segmentation clock as a model system. Our integration of experimental and theoretical approaches revealed that after desynchronization, the recovery of the iconic synchronized wave pattern in the segmentation clock is influenced by two distinct spatial and temporal scales. Firstly, there is the faster and local communication directly between the cells, and secondly, there is a slower and much longer-distance movement of the cells and the tissues in the embryo as the tissues change their overall shapes.
This work started about 10 years ago, when the four of us were all at the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) in Dresden, Germany. In fact, we all worked in the same research group, so local communication was relatively easy. This local discussion was triggered when one of us, Bo-Kai Liao, noticed an unusual pattern of disrupted body segments in zebrafish embryos as they attempted to re-synchronize their cellular clocks after perturbation. The thing that struck us was that the pattern really couldn’t be explained by the current models of re-synchronization.
We are sure you all recognize the situation: “Hang on a minute, that should not be happening…” and you just know you are going to find out something cool. Well, in this case, the journey from initial observation to an explanation would take us a decade, during which time all of us moved from the MPI-CBG. We each moved country at least once, and we changed jobs and titles and had families and even got some grey hair. The scientific story mirrored the personal: how local communication is affected by long-distance movements. To understand this story better, we need to go back to the beginning and start with the biology.
The precursors of the body segments of vertebrates, called somites, are formed rhythmically during embryonic development. Each segment buds off from the unsegmented tissue, presomitic mesoderm (PSM) one by one. The rhythm of segment formation is determined by an oscillatory spatial pattern of gene expression in the PSM and tailbud, termed the segmentation clock. In these tissues, the peaks of gene expression travel across the anterior-posterior axis. Cells in the tissue synchronize their gene expression rhythms with neighbors through Delta-Notch signaling, providing local integrity of gene expression patterns. Understanding this synchronization, and it’s role in forming body segments is what brought the four of us together in the first place.
Previous studies had shown that the treatment of an inhibitor of Delta-Notch signaling, called DAPT, led to formation of defective segments in zebrafish embryos. Some of these studies had also observed the recovery of normal segments after the washout of DAPT (Riedel-Kruse et al. 2007; Liao et al. 2016). These results had been interpreted in terms of desynchronization and resynchronization of oscillators: treatment of DAPT desynchronizes oscillators by inhibiting intercellular coupling through Delta-Notch signaling, and the hallmark spatial wave pattern of gene expression in the PSM is abolished due to the noise in the individual cellular oscillators. However, its washout restores coupling, letting cells gradually resynchronize their oscillators. When the synchrony level reaches a threshold, a normal segment reappears. This desynchronization hypothesis has quantitatively explained several experimental data, but there was a remaining gap in our understanding: how is the tissue-scale pattern reorganized through local coupling?
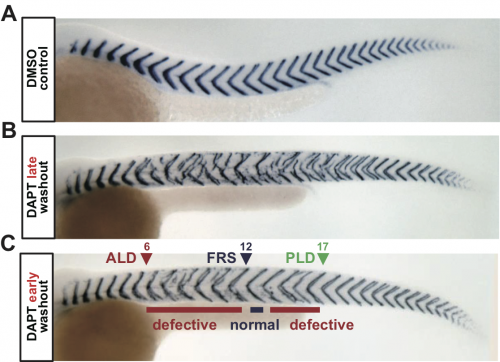
To address this, we analyzed segment recovery processes with different DAPT washout timing in zebrafish embryos. We unexpectedly found that washing out DAPT at earlier developmental stages caused intermingled segment boundary defects: a defective segment boundary was formed even after some normal segment boundaries were already formed (Fig. 1). This result suggests that the synchrony level fluctuates around the threshold, for some reason. We were very surprised by this result because we expected a monotonic recovery of normal segments based on previous theoretical works on coupled oscillators. Usually, once a population of oscillators gets synchronized, they remain synchronized and a large fluctuation of synchrony level hardly ever occurs. Thus, pattern recovery in the segmenting tissue seemed not so simple.
At this time, there were no transgenic markers of the zebrafish segmentation clock, nor were the microscopes, image processing, and data analysis developed to follow all these oscillating cells during the entire re-synch process (in fact, we are still working on this…).
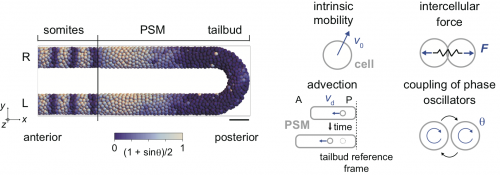
Therefore, we decided to use a physical model for the segmenting tissue to analyze this phenomenon. We have previously proposed models for the entire presomitic mesoderm and tailbud tissues in lower spatial dimension (1D or 2D; Morelli et al. 2009) or in 3D space but only a part of the tissue (Uriu et al. 2017). This time, we chose to describe the entire tissue in 3D and tried to integrate some of the previous modeling efforts by us and others in this framework (Fig. 2).
In numerical simulations of the model, we found a rotating phase pattern of oscillators, termed a phase vortex, in some situations of resynchronization (Fig. 3A, B). A phase vortex emerges in the tissue by local interactions of oscillators, moves through the tissue along the anterior-posterior axis by cell advection, and generates a defective boundary when it arrives at the anterior part of the PSM. A phase vortex can be formed posterior to the well-synchronized domain in simulation, so it can cause intermingled defective segments, as observed in the experiments.
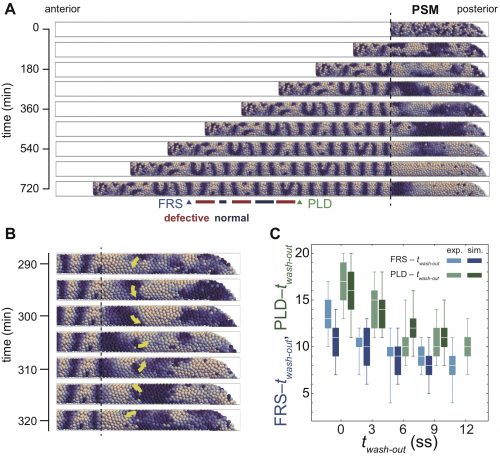
It turns out that although the formation of a phase vortex is driven by the local interactions, its kinematics is determined by the larger-scale tissue properties. A phase vortex moves from posterior to anterior by cell advection caused by embryonic axis extension. We reasoned that the global tissue properties, such as cell advection and tissue length could affect the pattern recovery in the PSM. Moreover, these tissue properties change with developmental stages. Hence, our physical model predicted that the time to complete recovery, that is, when we do not see any further phase vortices, would depend on the developmental stage at which DAPT washed out. In fact, we observed good agreement between simulations and experiment for the time to complete recovery (Fig. 3C). So, are we done? No. The next key task is to test the theory by observing phase vortices in living tissues. This should be possible by the live imaging of reporters of both oscillatory protein and segment boundaries simultaneously, which requires the long-term imaging, tracking and analysis of the cells in the segmentation clock.
In summary, our study indicated that pattern recovery in the zebrafish segmentation clock occurs at two spatial and temporal scales: quick local synchronization and transport of local patterns through slow tissue shape changes. There’s a nice symmetry to the slow, long-range drift of the four authors, which certainly changed the dynamics of our local communication. Although we were already quite good at video conferencing when the pandemic struck, the available time interval when everyone on the team was awake and alert simultaneously in Japan, Taiwan, Switzerland and Argentina was rather short. Thus, a high signaling strength was an important asset. Indeed, working together on the project over such a long time and through so many life changes was a rewarding experience, and we hope you will enjoy the pattern that self-organized from the process as much as we do.
Uriu K., Liao BK., Oates A.C., Morelli L.G. (2021) From local resynchronization to global pattern recovery in the zebrafish segmentation clock. Elife 10:e61358. doi: 10.7554/eLife.61358.
References
Liao BK., Jörg D.J., Oates A.C. (2016) Faster embryonic segmentation through elevated Delta-Notch signalling. Nat Commun. 7:11861. doi: 10.1038/ncomms11861.
Riedel-Kruse I.H., Müller C., Oates A.C. (2007) Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science 317(5846):1911-5. doi: 10.1126/science.1142538.
Morelli L.G., Ares S., Herrgen L., Schröter C., Jülicher F., Oates A.C. (2009) Delayed coupling theory of vertebrate segmentation. HFSP J. 3(1):55-66. doi: 10.2976/1.3027088.
Uriu K., Bhavna R., Oates A.C., Morelli L.G. (2017) A framework for quantification and physical modeling of cell mixing applied to oscillator synchronization in vertebrate somitogenesis. Biol Open. 6(8):1235-1244. doi: 10.1242/bio.025148.


 (No Ratings Yet)
(No Ratings Yet)