How we learned to build a gliding mammal
Posted by Jorge Moreno, on 2 September 2024
A new hope to study convergent evolution
Convergent evolution — the independent emergence of analogous structures among species whose last common ancestors lacked the trait— has long fascinated me. This phenomenon represents an exciting opportunity to study the genomic constraints that shape organisms during development to produce specific forms and functions. In the fall of 2018, as I was searching for a PhD program, I heard a talk by Dr. Ricardo Mallarino. He was interested in studying one such example of convergence, the evolution of the patagium, a skin membrane that, like a wingsuit, allows animals to glide through the air as a means of locomotion. Of particular interest was the patagium of a small marsupial possum: sugar gliders. The patagium has evolved many times across the tree of life including rodents, primates, marsupials, lizards, and frogs. Marsupials however, offered a uniquely tractable group to study how similar novel morphological traits evolved independently, as three closely related possums had acquired this adaptation. Here I will cover the findings of our recent publication.

Biology strikes back: identifying candidate regulatory elements
Previous RNAseq-based exploration of the developing patagium indicated that a major difference between the early patagium and neighboring skin was the differential regulation of an early gene regulatory network1. Given this, I set out to identify cis-regulatory elements (CREs) which had similar patterns of evolution in gliding species, which may harbor the causative changes driving differential gene regulation, evidenced by previous work2,3. We produced 15 new marsupial genomes which included gliding (sugar glider, greater glider, and feathertail glider) and non-gliding species. To identify cis-regulatory elements in the developing patagium tissue, we used ChIP- and ATAC-seq, which when used in combination provided a candidate list of active and poised CREs. Then using both our biological data and our genomes, we measured the rate of nucleotide change across these identified cis-regulatory elements, giving us an indication of which CREs are experiencing selection as these species have evolved their patagium. Through this analysis, we identified thousands of candidate glider accelerated regions (GARs)— elements which showed a substantial increase in nucleotide substitution. By focusing only on the GARs that showed shared patterns across the three gliding species we analyzed, we hoped to find a smaller pool of CREs that could be involved in the evolution of the patagium. We were surprised to find that not a single CRE matched that description. Then in our darkest times, a moment of brilliance: what if the cause was not a single CRE but instead different CREs across species all regulating the same gene. We had previously conducted Micro-C on the developing patagium and thought to use this data and identify topologically associating domains (TADs) to inform an analysis of GAR distribution and abundance. Using these TADs, we assigned CREs to the genes that were in the same TADs and asked if any gene had an overabundance of GARs.
Return of a key gene
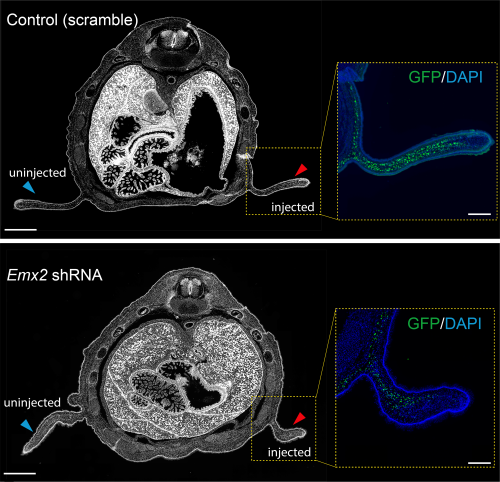
Previous work from our group uncovered that the sugar glider patagium develops through the deployment of a conserved network of genes, and that Wnt5a is heavily involved in the early development of the gliding membrane1. One of the other genes identified was the transcription factor Emx2. It just so happened, that our analysis for GAR enrichment identified Emx2 as our strongest candidate, having GARs from each of the three gliding species in its vicinity. We produced an shRNA lentivirus for Emx2 and began testing the effects of downregulating Emx2 in the developing patagium. Taking advantage of marsupial biology, that is they give birth to their young, or joeys, quite early in their development, we could then probe the early patagium. We conducted injections into the patagium primordium of sugar glider joeys and found that indeed downregulation of Emx2 caused a decrease in the area of the developing patagium. This was one of my favorite experiments and served to remind me how fascinating working in science can be. For some time, I was one of the only people in the world who knew that lack of Emx2 led to incorrect patagium growth. We then explored how Emx2 may be regulated. We had several candidates to test but initially focused on just two, one was positioned in what we presumed to be the promoter of Emx2 while the second seemed to be a distal enhancer located 1mb away from the Emx2 promoter but with a strong contact loop in the Micro-C dataset. The former was accelerated in the sugar glider and the latter accelerated in the feathertail glider. We wanted to test if the acceleration observed had an effect on the element’s ability to regulate expression and so we decided to use luciferase assays in an immortalized sugar glider cell line. This experiment works by placing your CRE of interest upstream of the luciferase gene (which originates from fireflies) to measure the amount of fluorescence produced by the cells to see if your CRE has regulatory function. In our case, our results indicated that the distal enhancer had accumulated changes that made it a stronger enhancer in the feathertail glider compared to its non-gliding sister and the sugar glider. We later found that the other gliding species, the greater glider, also had an enhancer that showed the same pattern.
Duo of fate: Our two favorite genes are important for patagium development
Now we knew that Emx2 had a phenotypic effect on the patagium and had some clues as to how it could end up being highly expressed in gliding species, but I became interested in what was happening at the molecular level in the patagium when we disrupted Emx2 expression. We did another round of shRNA injections, and this time collected the tissue for RNA sequencing. We found that many of the genes that were normally upregulated in the patagium when compared to surrounding skin were now downregulated. Among the ~400 genes downregulated, Wnt5a was one of the more strongly affected genes. This prompted us to investigate if Emx2 regulates all these genes directly and specifically how it may be regulating Wnt5a. We did an Emx2-ChIP-seq experiment and found that many genes did indeed have Emx2 binding sites, and we were able to identify multiple binding sites in the Wnt5a promoter. An example of great peer reviewing led to another of my favorite experiments as we set out to test the Emx2 binding sites found in the Wnt5a promoter. We again used luciferase this time testing two versions of the same promoter, one was unaltered while in the second we mutated the Emx2 binding sites. We found that the loss of these sites led to a complete loss of regulatory ability. Then to see if Emx2 was responsible for activating this promoter we co-transfected an Emx2-producing plasmid with our luciferase reporter plasmids. This experiment showed that the wild-type promoter increased its production of luciferase nearly 4-fold when Emx2 was present.
Emx2 awakens a conserved pathway
Our final goal for the paper was to establish if the spatial expression and function of Emx2 was novel to sugar gliders or if it was conserved in non-gliding mammals. We decided to test this hypothesis in mice, a much more amenable system for testing overexpression of genes. We found that Emx2, as reported previously4, was expressed in mice in a similar spatial pattern as that of the patagium, however this expression was only present for ~2 days whereas in the developing sugar gliders it was present for at least 14 days. To test whether overexpression of emx2 was sufficient to produce early patagium phenotypes, like we had previously observed with Wnt5, we extended the duration of Emx2 expression in mice while maintaining its endogenous spatial pattern. This however resulted in mice whose forebrain grew uncontrollably and resulted in non-viability; previous work had implicated Emx2 in brain development5. Therefore, we restricted the overexpression of Emx2 to only the skin. We found that indeed this overexpression was capable of recapitulating phenotypes observed in the early patagium such as increased cell proliferation, density, and the thickening of the epidermis1. These experiments showed that Emx2 has a conserved role in driving proliferation, potentially via regulation of the Wnt pathway, further indicating that evolution has re-used existing cellular programs to evolve a new adaptation.
The last remarks
In my opinion, the key message of this paper is that the evolution of convergent traits can occur independently via similar pathways/mechanisms, but the path to get there can be different. Our work showed that the redeployment of a shared developmental pathway can be an effective mechanism by which adaptations evolve.
I am very happy to see this work published; it took many years to get to this point. There were many bumps along the way, and it was the culmination of the hard work of many people involved. I want to thank the reviewers for their candid and helpful words, truly they made the paper better than when we first submitted. As someone who finds great joy in working on emerging model systems and has received countless advice in the past to just work on “model” organisms, I am incredibly pleased with how this paper is received. I continue to work on non-model systems with fascinating biology now as a postdoc and advise anyone who is interested in working on new models to go for it! It is rewarding to work on questions that can only be asked in a new system and uncover and share new findings.
References
1 Feigin, C. Y. et al. Convergent deployment of ancestral functions during the evolution of mammalian flight membranes. Science Advances 9, eade7511 (2023). https://doi.org/doi:10.1126/sciadv.ade7511
2 Booker, B. M. et al. Bat Accelerated Regions Identify a Bat Forelimb Specific Enhancer in the HoxD Locus. PLOS Genetics 12, e1005738 (2016). https://doi.org/10.1371/journal.pgen.1005738
3 Capra, J. A., Erwin, G. D., McKinsey, G., Rubenstein, J. L. R. & Pollard, K. S. Many human accelerated regions are developmental enhancers. Philosophical Transactions of the Royal Society B: Biological Sciences 368, 20130025 (2013). https://doi.org/doi:10.1098/rstb.2013.0025
4 Pellegrini, M., Pantano, S., Fumi, M. P., Lucchini, F. & Forabosco, A. Agenesis of the Scapula in Emx2 Homozygous Mutants. Developmental Biology 232, 149-156 (2001). https://doi.org/https://doi.org/10.1006/dbio.2001.0159
5 Yoshida, M. et al. Emx1 and Emx2 functions in development of dorsal telencephalon. Development 124, 101-111 (1997). https://doi.org/10.1242/dev.124.1.101


 (3 votes)
(3 votes)