July in preprints
Posted by the Node, on 1 August 2017
Our latest monthly trawl for developmental biology (and other cool) preprints. See last year’s introductory post for background, and let us know if we missed anything
This month we found lots of plant preprints and plenty of evo-devo work, as well as a series of preprints characterising features of the genome at distinct stages or places during development. The preprints were hosted on bioRxiv, PeerJ and arXiv. Use these links to get to the section you want:
| Stem cells, regeneration & disease modelling
Plants
Evo-devo & evo
Cell biology
Tools & resources
| Imaging etc.
Research practice
Why not…
Developmental biology
| Patterning & signalling
Drosophila embryonic type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. Kathleen T. Walsh, Chris Q. Doe
The RacGAP βChimaerin is essential for cerebellar granule cell migration. Jason A. Estep, Wenny Wong, Yiu-Cheung E. Wong, Brian M. Loui, Martin M. Riccomagno
PFKFB4 control of Akt signaling is essential for premigratory and migratory neural crest formation. Ana Leonor Figueiredo, Frederique Maczkowiak, Caroline Borday, Patrick Pla, Meghane Sittewelle, Caterina Pegoraro, Anne H. Monsoro-Burq
Exosomes regulate Neurogenesis and Circuit Assembly in a Model of Rett Syndrome. Pranav Sharma, Pinar Mesci, Cassiano Carromeu, Daniel McClatchy, Lucio Schiapparelli, John Yates, Alysson Muotri, Hollis T. Cline
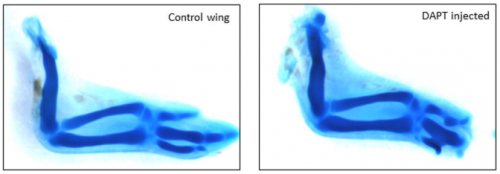
Hes1 oscillations synchronize and refine condensation formation and patterning of the avian limb skeleton. Ramray Bhat, Tilmann Glimm, Marta Linde-Medina, Cheng Cui, Stuart Newman
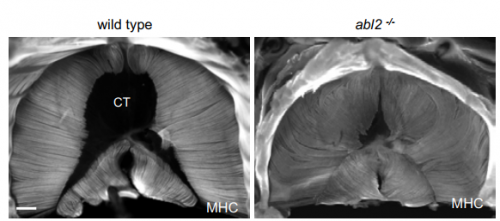
Abelson tyrosine-protein kinase 2 Regulates Myoblast Proliferation and Controls Muscle Fiber Length. Steven J Burden, Jennifer K Lee
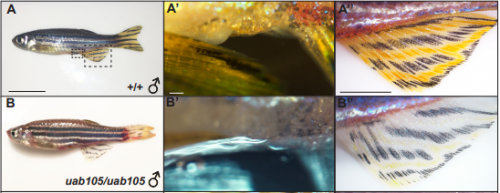
Nuclear androgen receptor regulates testes organization and oocyte maturation in zebrafish. Camerron M. Crowder, Christopher S. Lassiter, Daniel A. Gorelick
Three-dimensional cell neighbourhood impacts differentiation in the inner mass cells of the mouse blastocyst. Sabine C. Fischer, Elena Corujo-Simon, Joaquin Lilao-Garzon, Ernst H. K. Stelzer, Silvia Munoz-Descalzo
Fibronectin type III and intracellular domains of Toll-like receptor 4 interactor with leucine-rich repeats (Tril) are required for developmental signaling. Hyung-Seok Kim, Autumn McKnite, Yuanyuan Xie, Jan L. Christian
Progesterone regulates hypothalamic-pituitary-thyroid axis. Chunyun Zhong, Kewen Xiong, Xin Wang
Drosophila Kruppel homolog 1 represses lipolysis through interaction with dFOXO. Ping Kang, Kai Chang, Ying Liu, Mark Bousk, Galina Karashchuk, Rachel Thakore, Wenjing Zheng, Stephanie Post, Colin Brent, Sheng Li, Marc Tatar, Hua Bai
Non-cell autonomous Otx2 homeoprotein regulates visual cortex plasticity through Gadd45b. Jessica Apulei, Namsuk Kim, Damien Testa, Jérôme Ribot, Clémence Bernard, Ariel A. Di Nardo, Alain Prochiantz
| Morphogenesis & mechanics
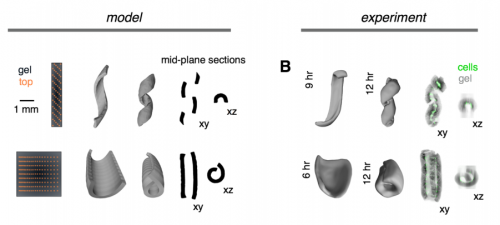
Tissue Folding by Mechanical Compaction of the Mesenchyme. Alex J. Hughes, Hikaru Miyazaki, Maxwell C. Coyle, Jesse Zhang, Matthew T. Laurie, Daniel Chu, Zuzana Vavrusova, Richard A. Schneider, Ophir D. Klein, Zev J. Gartner
Rap1 acts via multiple mechanisms to position Canoe/Afadin and adherens junctions and mediate apical-basal polarity establishment. Teresa Bonello, Kia Perez-Vale, Kaelyn Sumigray, Mark Peifer
A preferred curvature-based continuum mechanics framework for modeling embryogenesis with application to Drosophila mesoderm invagination. Khaled Khairy, William Lemon, Fernando Amat, Philipp Keller
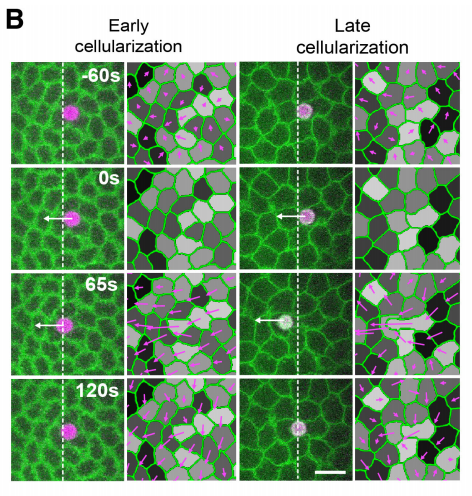
Probing tissue-scale deformation by in vivo force application reveals a fast tissue softening during early embryogenesis. Arturo D’Angelo, Kai Dierkes, Carlo Carolis, Guillaume Salbreux, Jerome Solon
Dystroglycan Maintains Inner Limiting Membrane Integrity To Coordinate Retinal Development. Reena Clements, Rolf Turk, Kevin Campbell, Kevin M. Wright
Feedback regulation of steady-state epithelial turnover and organ size. Jackson Liang, Shruthi Balachandra, Sang Ngo, Lucy Erin O’Brien
Image velocimetry and spectral analysis enable quantitative characterization of larval zebrafish gut motility. Julia Ganz, Ryan P. Baker, M. Kristina Hamilton, Ellie Melancon, Parham Diba, Judith S. Eisen, Raghuveer Parthasarathy
The ABC transporter Snu drives formation of the lipid-based inward and outward barrier in the skin of Drosophila. Renata Zuber, Michaela Norum, Yiwen Wang, Kathrin Oehl, Davide Accardi, Slawomir Bartoszewski, Nicole Gehring, Jurgen Berger, Matthias Flotenmeyer, Bernard Moussian
Exploring the underlying mechanisms of Xenopus laevis embryonic cell cycle. Kun Zhang, Jin Wang.
Roles of Diffusion Dynamics and Molecular Concentration Gradients in Cellular Differentiation and Three-Dimensional Tissue Development. Richard J. McMurtrey
Effects of developmental variability on the dynamics and self-organization of cell populations. Kaumudi H Prabhakara, Azam Gholami, Vladimir S Zykov, Eberhard Bodenschatz
| Genes & genomes
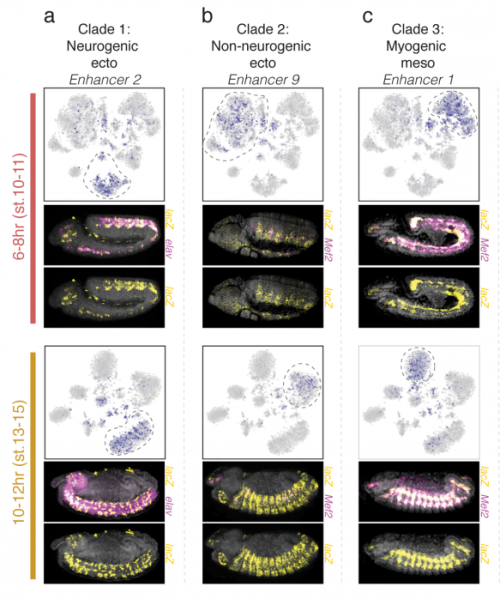
The cis-regulatory dynamics of embryonic development at single cell resolution. Darren A. Cusanovich, James P. Reddington, David A. Garfield, Riza Daza, Raquel Marco-Ferreres, Lena Christiansen, Xiaojie Qiu, Frank Steemers, Cole Trapnell, Jay Shendure, Eileen E.M. Furlong
Transcription start site profiling uncovers divergent transcription and enhancer-associated RNAs in Drosophila melanogaster. Michael P. Meers, Karen Adelman, Robert J Duronio, Brian D Strahl, Daniel J McKay, A. Gregory Matera
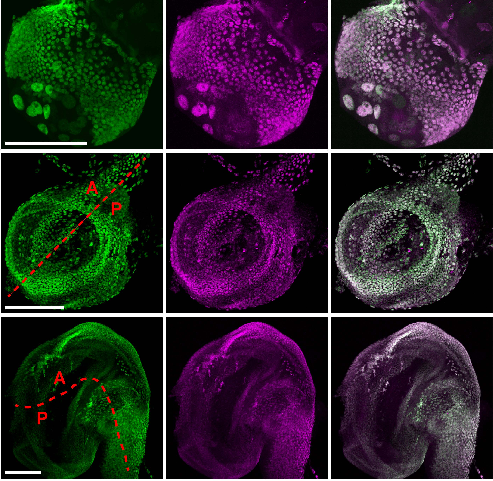
Control of Hox transcription factor concentration and cell-to-cell variability by an auto-regulatory switch. Dimitrios K. Papadopoulos, Kassiani Skouloudaki, Ylva Engstrom, Lars Terenius, Rudolf Rigler, Christoph Zechner, Vladana Vukojevic, Pavel Tomancak
The TAF10-containing TFIID and SAGA transcriptional complexes are dispensable for early somitogenesis in the mouse embryo. Paul Bardot, Stephane D. Vincent, Marjorie Fournier, Alexis Hubaud, Mathilde Joint, Laszlo Tora, Olivier Pourquie
BRG1/BRM-associated factor complex subunit diversity promotes temporally distinct gene expression programs in cardiogenesis. Swetansu Hota, Jeffrey Johnson, Erik Verschueren, Yiwen Zhu, Xin Sun, Len Pennacchio, Janet Rossant, Nevan Krogan, Benoit Bruneau
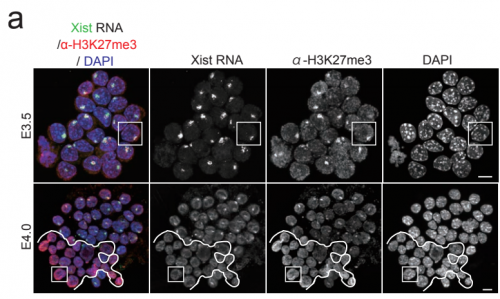
Differential epigenetic landscapes and transcription factors explain X-linked gene behaviours during X-chromosome reactivation in the mouse inner cell mass. Maud Borensztein, Ikuhiro Okamoto, Laurene Syx, Guillaume Guilbaud, Christel Picard, Katia Ancelin, Rafael Galupa, Patricia Dianbangouaya, Nicolas Servant, Emmanuel Barillot, Azim Surani, Mitinori Saitou, Chong-Jian Chen, Konstantinos Anastassiadis, Edith Heard
Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome.Giancarlo Bonora, Xinxian Deng, He Fang, Vijay Ramani, Ruolan Qui, Joel Berletch, Gala N Filippova, Zhijun Duan, Jay Schendure, William S Noble, Christine M Disteche
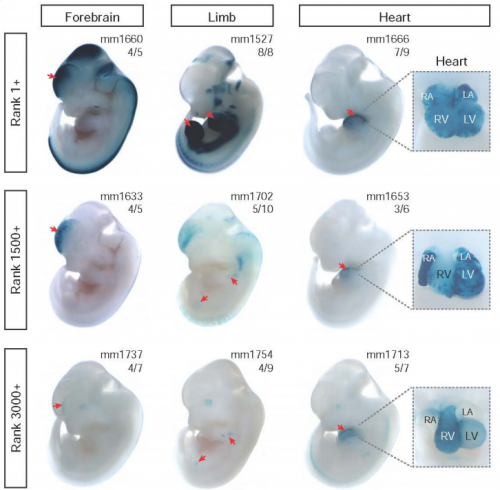
Systematic mapping of chromatin state landscapes during mouse development. David Gorkin, Iros Barozzi, Yanxiao Zhang, Ah Young Lee, Bin Lee, Yuan Zhao, Andre Wildberg, Bo Ding, Bo Zhang, Mengchi Wang, J. Seth Strattan, Jean M Davidson, Yunjiang Qiu, Veena Afzal, Jennifer A Akiyama, Ingrid Plajzer-Frick, Catherine S Pickle, Momoe Kato, Tyler H Garvin, Quan T Pham, Anne N Harrington, Brandon J Mannion, Elizabeth A Lee, Yoko Fukuda-Yuzawa, Yupeng He, Sebastian Preissl,Sora Chee, Brian A Williams, Diane Trout, Henry Amrhein, Hongbo Yang, J. Michael Cherry, Yin Shen, Joseph R Ecker, Wei Wang, Diane E Dickel, Axel Visel, Len A Pennacchio, Bing Ren
Systems-level identification of transcription factors critical for mouse embryonic development. Kai Zhang, Mengchi Wang, Ying Zhao, Wei Wang
Nonparametric Bayesian inference of transcriptional branching and recombination identifies regulators of early human germ cell development. Christopher Andrew Penfold, Anastasiya Sybirna, John Reid, Yun Huang, Lorenz Wernisch, Zoubin Ghahramani, Murray Grant, M. Azim Surani
Specification of the germline by Nanos-dependent down-regulation of the somatic synMuvB transcription factor LIN-15B. Chih-Yung Sean Lee, Tu Lu, Geraldine Seydoux
Spatiotemporal DNA Methylome Dynamics of the Developing Mammalian Fetus. Yupeng He, Manoj Hariharan, David U Gorkin, Diane E Dickel, Chongyuan Luo, Rosa G Castanon, Joseph R Nery, Ah Young Lee, Brian A Williams, Diane Trout, Henry Amrhein,Rongxin Fang, Huaming Chen, Bin Li, Axel Visel, Len A Pennacchio, Bing Ren, Joseph R Ecker
Single nucleus analysis of the chromatin landscape in mouse forebrain development. Sebastian Preissl, Rongxin Fang, Yuan Zhao, Ramya Raviram, Yanxiao Zhang, Brandon C. Sos, Hui Huang, David U. Gorkin, Veena Afzal, Diane E. Dickel, Samantha Kuan, Axel Visel, Len A. Pennacchio, Kun Zhang, Bing Ren
Quantitative RNAseq Meta Analysis Of Alternative Exon Usage In C. elegans. Nicolas J. Tourasse, Jonathan R. M. Millet, Denis Dupuy
Genetic variation and gene expression across multiple tissues and developmental stages in a non-human primate. Anna J. Jasinska, Ivette Zelaya, Susan K. Service, Christine Peterson, Rita M. Cantor, Oi-Wa Choi, Joseph DeYoung, Eleazar Eskin, Lynn A. Fairbanks, Scott Fears, Allison Furterer, Yu S. Huang, Vasily Ramensky, Christopher A. Schmitt, Hannes Svardal, Matthew J. Jorgensen, Jay R. Kaplan, Diego Villar, Bronwen L. Aken, Paul Flicek, Rishi Nag, Emily S. Wong, John Blangero, Thomas D. Dyer, Marina Bogomolov, Yoav Benjamini, George M. Weinstock, Ken Dewar, Chiara Sabatti, Richard K. Wilson, J. David Jentsch, Wesley Warren, Giovanni Coppola, Roger P. Woods, Nelson B. Freimer
Interplay of cis and trans mechanisms driving transcription factor binding and gene expression evolution. Emily S. Wong, Bianca M. Schmitt, Anastasiya Kazachenka, David Thybert, Aisling Redmond, Frances Connor, Tim F. Rayner, Christine Feig, Anne C. Ferguson-Smith, John C. Marioni, Duncan T. Odom, Paul Flicek
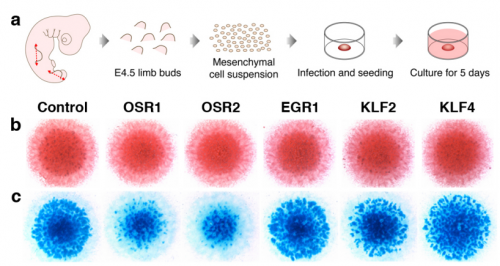
Genome-wide strategies identify molecular niches regulated by connective tissue-associated transcription factors. Mickael Orgeur, Marvin Martens, Georgeta Leonte, Sonya Nassari, Marie-Ange Bonnin, Stefan T Börno, Bernd Timmermann, Jochen Hecht, Delphine Duprez, Sigmar Stricker
Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Elizaveta Radion, Valeriya Morgunova, Sergei Ryazansky, Natalia Akulenko, Sergey Lavrov, Yuri Abramov, Ivan Olovnikov, Alla Kalmykova
The genetics of the human face: identification of large effect single gene variants. Daniel JM Crouch, Bruce Winney, Willem P Koppen, William J Christmas, Katarzyna Hutnik, Tammy Day, Devendra Meena, Abdelhamid Boumertit, Pirro Hysi, Ayrun Nessa, Tim D Spector, Josef Kittler, Walter F Bodmer
Sub-kb resolution Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Qi Wang, Qiu Sun, Daniel M. Czajkowsky, Zhifeng Shao
| Stem cells, regeneration & disease modelling
In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Alyssa J. Miller, David R. Hill, Melinda S. Nagy, Yoshiro Aoki, Briana R. Dye, Alana M. Chin, Sha Huang, Felix Zhu, Eric S. White, Vibha Lama, Jason R. Spence
Functional dissection of the enhancer repertoire in human embryonic stem cells. Tahsin Stefan Barakat, Florian Halbritter, Man Zhang, Andre Figueiredo Rendeiro, Christoph Bock, Ian Chambers
The transcriptionally permissive chromatin state of ES cells is acutely tuned to translational output. Aydan Bulut-Karslioglu, Trisha A Macrae, Juan A Oses-Prieto, Sergio Covarrubias, Michelle Percharde, Gregory Ku, Aaron Diaz, Michael T McManus, Alma L Burlingame, Miguel Ramalho-santos
Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli are Altered with Age. Mati Mann, Arnav Mehta, Carl de Boer, Monika S. Kowalczyk, Kevin Lee, Daneyal Farouq, Aviv Regev, David Baltimore
Clonal Dominance and Transplantation Dynamics in Hematopoietic Stem Cell Compartments. Peter Ashcroft, Markus G. Manz, Sebastian Bonhoeffer
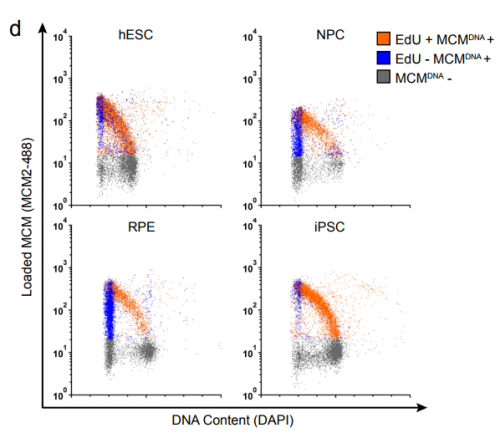
Rapid DNA Replication Origin Licensing Protects Stem Cell Pluripotency. Jacob Peter Matson, Raluca Dumitru, Phillip Coryell, Ryan M. Baxley, Weili Chen, Kirk Twaroski, Beau R. Webber, Jakub Tolar, Anja-Katrin Bielinsky, Jeremy Purvis, Jeanette Gowen Cook
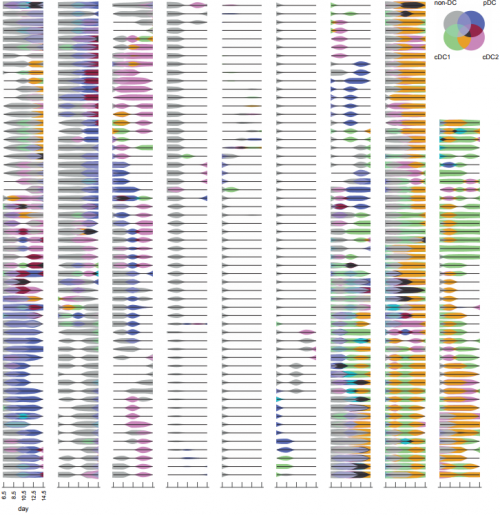
Assessment of clonal kinetics reveals multiple trajectories of dendritic cell development. Dawn Lin, Andrey Kan, Jerry Gao,Edmund Crampin, Philip D. Hodgkin, Shalin H. Naik
Dynamic expression of tRNA-derived small RNAs define cellular states. Daniel Yim, Srikar Krishna, Vairavan Lakshmanan, Judice Koh, Jung Eun Park, Jit Kong Cheong, Joo Leng Low, Michelle Lim, Junyu Ip, Jie Min Nah, Iain Tan, N. Gopalakrishna Iyer, Huili Guo, Siu Kwan Sze, Srikala Raghavan, Dasaradhi Palakodeti, Ramanuj DasGupta
Deep convolutional neural networks allow analysis of cell motility during stem cell differentiation and neoplastic transformation. Jacob Kimmel, Andrew Brack, Wallace F. Marshall
An Algorithm for Cellular Reprogramming. Scott Ronquist, Geoff Patterson, Markus Brown, Stephen Lindsly, Haiming Chen, Lindsey Muir, Max Wicha, Anthony Bloch, Roger Brockett, Indika Rajapakse
Recasting the cancer stem cell hypothesis: unification using a continuum model of microenvironmental forces. Jacob G. Scott, Andrew Dhawan, Anita Hjelmeland, Justin Lathia, Masahiro Hitomi, Alexander G. Fletcher, Philip K. Maini, Alexander R. A. Anderson
Suppression of Transposable Elements in Leukemic Stem Cells. Anthony Richard Colombo, Asif Zubair, Devi Thiagarajan, Sergey Nuzhdin, Timothy J. Triche Jr., Giridharan Ramsingh
Early postnatal brain overgrowth and gene expression changes prefigure functional over-connectivity of the cortex in Chd8 haploinsufficient mice. Philipp Suetterlin, Shaun Hurley, Conor Mohan, Kimberley L. H. Riegman, Marco Pagani, Angela Caruso, Jacob Ellegood, Alberto Galbusera, Ivan Crespo-Enriquez, Caterina Michetti, Robert Ellingford, Olivier Brock, Alessio Delogu, Philippa Francis-West, Jason P. Lerch, Maria Luisa Scattoni, Alessandro Gozzi, Cathy Fernandes, Albert Basson
Extracellular monomeric and aggregated tau efficiently enter human neurons through overlapping but distinct pathways. Thomas Wassmer, Lewis Evans, Graham Fraser, James Smith, Michael Perkinton, Andy Billinton, Frederick Livesey
Changes in extracellular matrix cause RPE cells to make basal deposits and activate the alternative complement pathway. Rosario Fernandez-Godino, Kinga M. Bujakowska, Eric A. Pierce
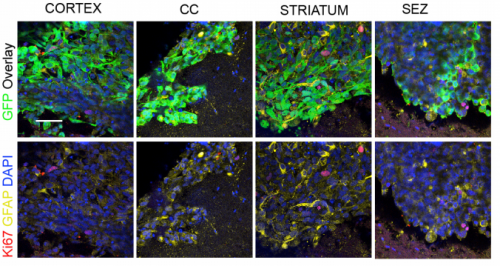
Modelling glioblastoma tumour-host cell interactions using adult brain organotypic slice co-culture. Maria Angeles Marques, Ester Gangoso, Steven Pollard
Assaying uptake of endocrine disruptor compounds in zebrafish embryos and larvae. Jaclyn Paige Souder, Daniel A. Gorelick
Plants
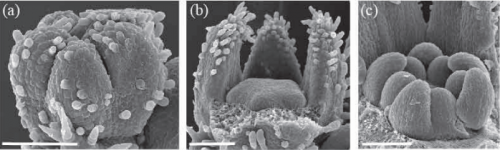
Heterochronic Developmental Shifts Underlie Floral Diversity within Jaltomata (Solanaceae). Jamie L. Kostyun, Jill C. Preston, Leonie C. Moyle
Rice TSV3 Encoding Obg-like GTPase Protein is Essential for Chloroplast Development during the Early Leaf Stage under Cold Stress. Dongzhi Lin, Quan Jiang, Xiaojing Ma, Kailun Zheng, Xiaodi Gong, Sheng Teng, Jianlong Xu, Yanjun Dong
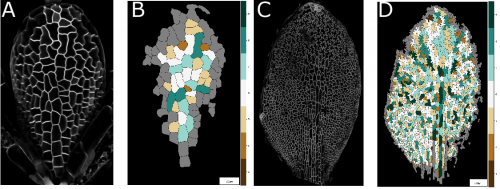
Pavement cells and the topology puzzle. Ross Carter, Yara Elena Sanchez-Corrales, Veronica A. Grieneisen, Athanasius F. M. Maree
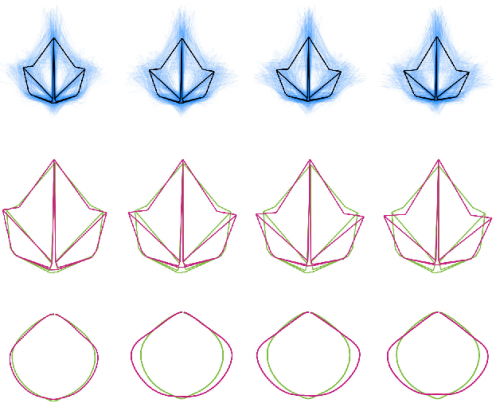
Divergent leaf shapes among Passiflora species arise from a shared juvenile morphology. Daniel H. Chitwood, Wagner C. Otoni
SANe: The Seed Active Network For Mining Transcriptional Regulatory Programs of Seed Development. Chirag Gupta, Arjun Krishnan, Eva Collakova, Pawel Wolinski, Andy Pereira
A transcription factor binding atlas for photosynthesis in cereals identifies a key role for coding sequence in the regulation of gene expression. Steven J. Burgess, Ivan Reyna-Llorens, Katja Jaeger, Julian M. Hibberd
Biomass increase under zinc deficiency caused by delay of early flowering in Arabidopsis. Xiaochao Chen, Uwe Ludewig
A Framework to Simulate Cortical microtubule Dynamics in Arbitrary Shaped Plant Cells. Bandan Chakrabortty, Ben Scheres, Bela Mulder
Pollen tube growth and guidance: Occam’s razor sharpened on a molecular AGP Rosetta Stone. Derek T. A. Lamport, Li Tan, Michael Held, Marcia J. Kieliszewski
Genetic Architecture and Molecular Networks Underlying Leaf Thickness in Desert-Adapted Tomato Solanum pennellii. Viktoriya Coneva, Margaret H. Frank, Maria A. de Luis Balaguer, Mao Li, Rosangela Sozzani, Daniel H. Chitwood
The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Zhaohe Yuan, Yanming Fang, Taikui Zhang, Zhangjun Fei, Fengming Han, Cuiyu Liu, Min Liu, Wei Xiao, Wenjing Zhang, Mengwei Zhang, Youhui Ju, Huili Xu, He Dai, Yujun Liu, Yanhui Chen, Lili Wang, Jianqing Zhou, Dian Guan, Ming Yan, Yanhua Xia, Xianbin Huang, Dongyuan Liu, Hongmin Wei, Hongkun Zheng
The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Aleksey V. Zimin, Daniela Puiu, Richard Hall, Sarah Kingan, Bernardo J. Clavijo,Steven L. Salzberg
Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Kelsey A. Maher, Marko Bajic, Kaisa Kajala, Mauricio Reynoso, Germain Pauluzzi, Donnelly A. West, Kristina Zumstein, Margaret Woodhouse, Kerry Bubb, Mike Dorrity, Christine Queitsch, Julia Bailey-Serres, Neelima Sinha, Siobhan M. Brady, Roger B. Deal
Epigenetic activation of meiotic recombination in Arabidopsis centromeres via loss of H3K9me2 and non-CG DNA methylation. Charles J. Underwood, Kyuha Choi, Christophe Lambing, Xiaohui Zhao, Heidi Serra, Filipe Borges, Joe Simorowski, Evan Ernst, Yannick Jacob, Ian R. Henderson, Robert A. Martienssen
Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis transposons and gene regulatory regions. Kyuha Choi, Xiaohui Zhao, Christophe Lambing, Charles J. Underwood, Thomas J. Hardcastle, Heidi Serra, Andrew J. Tock, Piotr A. Ziolkowski, Nataliya E. Yelina, Robert A. Martienssen, Ian R. Henderson
Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Heidi Serra, Christophe Lambing, Catherine H. Griffin, Stephanie D. Topp, Mathilde Seguela-Arnaud, Joiselle Fernandes, Raphael Mercier, Ian R. Henderson
Unleashing meiotic crossovers in hybrid plants. Joiselle B. Fernandes, Mathilde Seguela-Arnaud, Cecile Larcheveque, Andrew H. Lloyd, Raphael Mercier
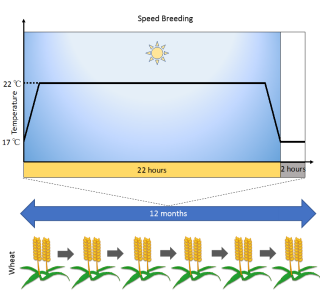
Speed breeding: a powerful tool to accelerate crop research and breeding. Amy Watson, Sreya Ghosh, Matthew Williams, William S. Cuddy, James Simmonds, Maria-Dolores Rey, M. Asyraf Md Hatta, Alison Hinchliffe, Andrew Steed, Daniel Reynolds, Nikolai Adamski, Andy Breakspear, Andrey Korolev, Tracey Rayner, Laura E. Dixon, Adnan Riaz, William Martin, Merrill Ryan, David Edwards, Jacqueline Batley, Harsh Raman, Christian Rogers, Claire Domoney, Graham Moore, Wendy Harwood, Paul Nicholson, Mark J. Dieters, Ian H. DeLacy, Ji Zhou, Cristobal Uauy, Scott A. Boden, Robert F. Park, Brande B. H. Wulff, Lee T. Hickey
Maize leaf functional responses to drought episode and rewatering. He Song, Yibo Li, Li Zhou, Zhenzhu Xu, Guangsheng Zhou
Evo-devo & evo
The origin of animal multicellularity and cell differentiation. Thibaut Brunet, Nicole King
Adaptive evolution of proteins expressed in late and post-embryonic development in animals. Jialin Liu, Marc Robinson-Rechavi
Developmental constraints on genome evolution in four bilaterian model species. Jialin Liu, Marc Robinson-Rechavi
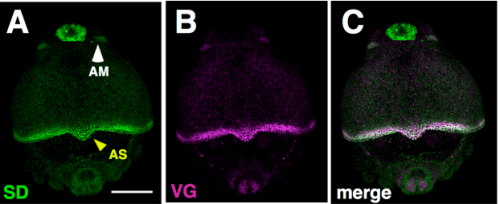
Repeated co-option of a conserved gene regulatory module underpins the evolution of the crustacean carapace, insect wings and other flat outgrowths. Yasuhiro Shiga, Yasuhiko Kato, Yuko Aragane-Nomura, Takayuki Haraguchi, Theodora Saridaki, Hajime Watanabe, Taisen Iguchi, Hideo Yamagata, Michalis Averof
Whole-body single-cell sequencing of the Platynereis larva reveals a subdivision into apical versus non-apical tissues. Kaia Achim, Nils Eling, Hernando Martinez Vergara, Paola Yanina Bertucci, Thibaut Brunet, Paul Collier, Vladimir Benes, John C Marioni, Detlev Arendt
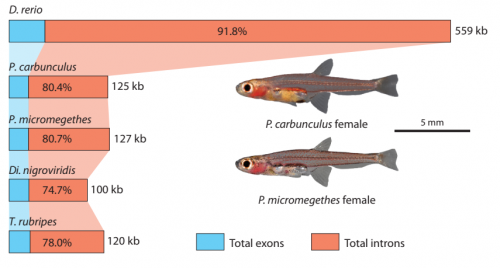
The most developmentally truncated fishes show extensive Hox gene loss and miniaturized genomes. Martin Malmstrøm, Ralf Britz, Michael Matschiner, Ole Kristian Tørresen, Renny Kurnia Hadiaty, Norsham Yaakob, Heok Hui Tan, Kjetill Sigurd Jakobsen, Walter Salzburger, Lukas Rüber
A novel flatworm-specific gene family implicated in reproduction in Macrostomum lignano. Magda Grudniewska, Stijn Mouton, Margriet Grelling, Anouk H. G. Wolters, Jeroen Kuipers, Ben N. G. Giepmans, Eugene Berezikov
Discovery of novel representatives of bilaterian neuropeptide families and reconstruction of neuropeptide precursor evolution in ophiuroid echinoderms. Meet Zandawala, Ismail Moghul, Luis Alfonso Yanez Guerra, Jerome Delroisse, Nikara Abylkassimova, Andrew F. Hugall, Timothy D. O’Hara, Maurice R. Elphick
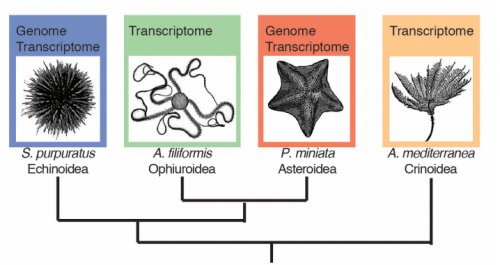
Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. David V Dylus, Liisa M Blowes, Anna Czarkwiani, Maurice R Elphick, Paola Oliveri
β-catenin has both conserved and novel functions in the sponge Ephydatia muelleri. Klaske J. Schippers, Scott A. Nichols
Ancestrally Shared Regenerative Mechanisms Across the Metazoa: A Transcriptomic Case Study in the Demosponge Halisarca caerulea. Nathan James Kenny, Jasper M. de Goeij, Didier M. de Bakker, Casey G. Whalen, Eugene Berezikov, Ana Riesgo
Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. David Thybert, Maša Roller, Fábio C. P. Navarro, Ian Fiddes, Ian Streeter, Christine Feig, David Martin-Galvez, Mikhail Kolmogorov, Václav Janoušek, Wasiu Akanni, Bronwen Aken, Sarah Aldridge, Varshith Chakrapani, William Chow, Laura Clarke, Carla Cummins, Anthony Doran, Matthew Dunn, Leo Goodstadt, Kerstin Howe, Matthew Howell, Ambre-Aurore Josselin, Robert C. Karn, Christina M. Laukaitis, Lilue Jingtao, Fergal Martin, Matthieu Muffato, Michael A. Quail, Cristina Sisu, Mario Stanke, Klara Stefflova, Cock Van Oosterhout, Frederic Veyrunes, Ben Ward, Fengtang Yang, Golbahar Yazdanifar, Amonida Zadissa, David Adams, Alvis Brazma, Mark Gerstein, Benedict Paten, Son Pham, Thomas Keane, Duncan T. Odom, Paul Flicek
Signatures of the evolution of parthenogenesis and cryptobiosis in the genomes of panagrolaimid nematodes. Philipp H. Schiffer, Etienne Danchin, Ann M. Burnell, Anne-Marike Schiffer, Chris Creevey, Simon Wong, Ilona Dix, Georgina O’Mahony, Bridget A. Culleton, Corinne Rancurel, Gary Stier, Elizabeth A. Martinez-Salazar, Aleksandra Marconi, Urmi Trivedi, Michael Kroiher, Michael A. S. Thorne, Einhard Schierenberg, Thomas Wiehe, Mark Blaxter
Divergence times in demosponges (Porifera): first insights from new mitogenomes and the inclusion of fossils in a birth-death clock model. Astrid Schuster, Sergio Vargas, Ingrid Knapp, Shirley A. Pomponi, Robert J. Toonen, Dirk Erpenbeck, Gert Woerheide
Fantastic beasts and how to sequence them: genomic approaches for obscure model organisms. Mikhail V Matz
Challenges and advances for transcriptome assembly in non-model species. Arnaud Ungaro, Nicolas Pech, Jean-Francois Martin, Scott RJ McCairns, Jean-Philippe Mevy, Remi Chappaz, Andre Gilles
Whole genome bisulfite sequencing reveals a sparse, but robust pattern of DNA methylation in the Dictyostelium discoideum genome. Jacob Steenwyk, James St. Denis, Jacqueline Dresch, Denis Larochelle, Robert Drewell

Phylogenomics illuminates the backbone of the Myriapoda Tree of Life and reconciles morphological and molecular phylogenies. Rosa Fernandez, Gregory D Edgecombe, Gonzalo Giribet
Multimodal Sensorimotor System in Unicellular Zoospores of a Fungus. Andrew Swafford, Todd Oakley
Evolutionary origins and diversification of testis-specific short histone H2A variants in mammals. Antoine Molaro, Janet M. Young, Harmit Singh Malik
Genome expansion and lineage-specific genetic innovations in the world’s largest organisms (Armillaria). Gyorgy Sipos, Arun N Prasanna, Mathias C Walther, Eoin O’Connor, Balazs Balint, Krisztina Krizsan, Brigitta Kiss, Jaqueline Hess, Jason Slot, Robert Riley, Bettina Boka, Daniel Rigling, Kerrie Barry, Juna Lee, Sirma Mihaltseva, Kurt Labutti, Anna Lipzen, Rose Waldron, Nicola Moloney, Christoph Sperisen, Laszlo Kredics, Csaba Vagvolgyi, Andrea Patrigniani, David Fitzpatrick, Istvan Nagy, Sean Doyle, James B Anderson, Igor V Grigoriev, Ulrich Guldener, Martin Munsterkotter, Torda Varga, Laszlo G Nagy
Cell biology
Drosophila PLP forms centriolar-clouds that promote centriole stability, cohesion and MT nucleation. Helio Roque, Metta Pratt, Errin Johnson, Jordan Raff
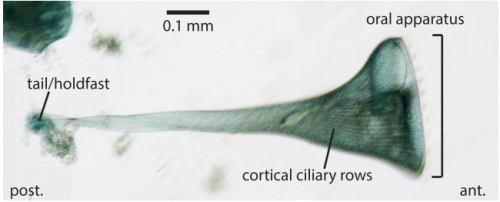
A large kinome in a large cell: Stentor coeruleus possesses highly expanded kinase families and novel domain architectures. Sarah B. Reiff, Wallace F. Marshall
Modest static pressure suppresses columnar epithelial cell proliferation in association with cell shape and cytoskeletal modifications. Man Hagiyama, Norikazu Yabuta, Daisuke Okuzaki, Takao Inoue, Yasutoshi Takashima, Ryuichiro Kimura, Aritoshi Ri, Akihiko Ito
Positive Feedback Between Contractile Ring Myosin and Ring-Directed Cortical Flow Drives Cytokinesis. Renat Khaliullin, Rebecca Green, Linda Shi, Michael Berns, J. Sebastian Gomez-Cavazos, Arshad Desai, Karen Oegema
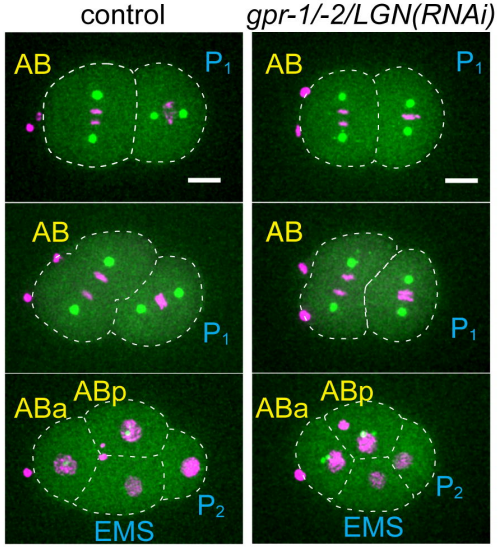
Cell-nonautonomously tunable actomyosin flows orient distinct cell division axes. Kenji Sugioka, Bruce Bowerman
The mitotic spindle is chiral due to torques generated by motor proteins. Maja Novak, Bruno Polak, Juraj Simunic, Zvonimir Boban, Barbara Kuzmic, Andreas Thomae, Iva M. Tolic, Nenad Pavin
Self-organization mechanism of distinct microtubule orientations in axons and dendrites. Honda Naoki, Katsuhiro Uegaki, Shin Ishii
C3G dynamically associates with Nuclear speckles and regulates mRNA splicing. Dhruv Kumar Shakyawar, Muralikrishna Bhattiprolu, Vegesna Radha
Context-dependent deposition and regulation of mRNAs in P-bodies. Congwei Wang, Fabian Schmich, Julie Weidner, Niko Beerenwinkel, Anne Spang
Loss Of PTEN Promotes Formation Of Signaling-Capable Clathrin-Coated Pits. Luciana K. Rosselli-Murai, Joel A. Yates, Sei Yoshida, Julia T. Bourg, Kenneth K. Y. Ho, Megan White, Julia Prisby, Xinyu Tan, Megan Altemus, Liwei Bao, Zhi-Fen Wu, Sarah L. Veatch, Joel A. Swanson, Sofia D. Merajver, Allen P. Liu
lncRNA H19 mediates TGF-β1-induced epithelial to mesenchymal transition in bovine epithelial cells through PI3K/AKT Signaling Pathway. wei yang, xuezhong li, shaopei qi, xueru li, kun zhou, suzhu qing, yong zhang, Mingqing Gao
Tools & resources
| Imaging etc.
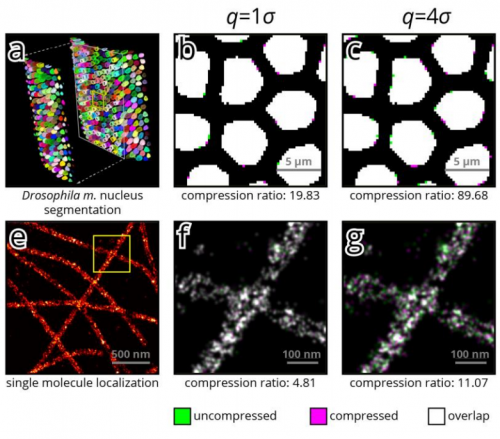
A real-time compression library for microscopy images. Balint Balazs, Joran Deschamps, Marvin Albert, Jonas Ries, Lars Hufnagel
NanoJ-SQUIRREL: quantitative mapping and minimisation of super-resolution optical imaging artefacts. Siân Culley, David Albrecht, Caron Jacobs, Pedro Matos Pereira, Christophe Leterrier, Jason Mercer, Ricardo Henriques
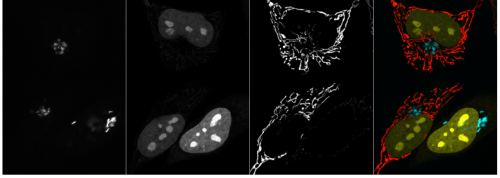
Robust and Bright Genetically Encoded Fluorescent Markers for Highlighting Structures and Compartments in Mammalian Cells. Anna O Chertkova, Marieke Mastop, Marten Postma, Nikki van Bommel, Sanne van der Niet, Kevin L. Batenburg, Linda Joosen, Theodorus W.J. Gadella Jr., Yasushi Okada, Joachim Goedhart
Improving Phenotypic Measurements in High-Content Imaging Screens. D. Michael Ando, Cory McLean, Marc Berndl
RAPID: Real-time image-based autofocus for all wide-field optical microscopy systems. Ludovico Silvestri, Marie Caroline Muellenbroich, Irene Costantini, Antonino Paolo Di Giovanna, Leonardo Sacconi, Francesco Saverio Pavone
Advances And Perspectives In Tissue Clearing Using CLARITY. Kristian H. R. Jensen,Rune W. Berg
Monomerization of Far-Red Fluorescent Proteins. Timothy M Wannier, Sarah Gillespie, Nicholas Hutchins, R Scott McIsaac, Kevin S Brown, Stephen L Mayo
A Fluorogenic Array Tag for Temporally Unlimited Single Molecule Tracking. Rajarshi P Ghosh, Matthew J Franklin, Will E Draper, Quanming Shi, Jan T Liphardt
A 340/380 nm Light Emitting Diode Illuminator For Fura-2 AM Ratiometric Ca2+ Imaging Of Live Cells With Better Than 5 nM Precision. Peter W. Tinning, Aimee J. P.M. Franssen, Shehla U. Hridi, Trevor J. Bushell, Gail McConnell
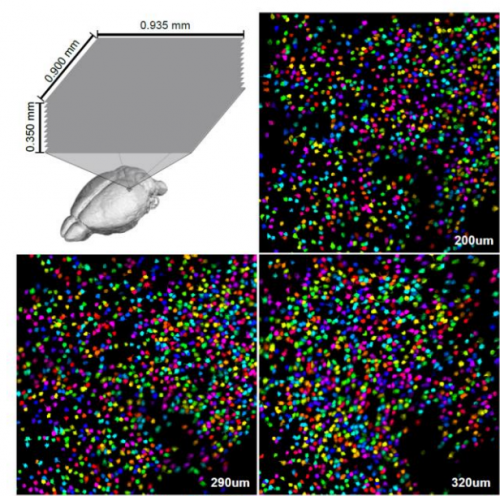
Suite2p: beyond 10,000 neurons with standard two-photon microscopy. Marius Pachitariu, Carsen Stringer, Mario Dipoppa, Sylvia Schröder, L. Federico Rossi, Henry Dalgleish, Matteo Carandini, Kenneth D Harris
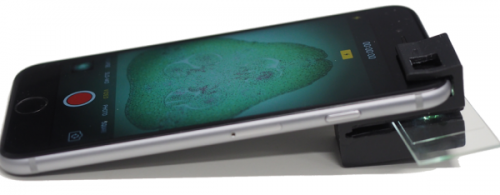
A dual-mode mobile phone microscope using integrated and ambient light. Antony Orth, Emma Wilson, Jeremy Thompson, Brant C. Gibson
Versatile open software to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Luca Sala, Berend J. van Meer, Leon G. J. Tertoolen, Jeroen Bakkers, Milena Bellin, Richard Davis, Chris Denning, Michel A. E. Dieben, Thomas Eschenhagen, Elisa Giacomelli, Catarina Grandela, Arne Hansen, Eduard R. Holman, Monique R. M. Jongbloed, Sarah M. Kamel, Charlotte D. Koopman, Quentin Lachaud, Ingra Mannhardt, Mervyn P. H. Mol, Valeria V. Orlova, Robert Passier, Marcelo C. Ribeiro, Umber Saleem, Godfrey L. Smith, Christine L. Mummery, Francis L. Burton
ShapeRotator: an R package for standardised rigid rotations of articulated Three-Dimensional structures with application for geometric morphometrics. Marta Vidal-Garcia, Lashi Bandara, J. Scott Keogh
CRISPR/Cas9-mediated Knock-in of an Optimized TetO Repeat for Live Cell Imaging of Endogenous Loci. Ipek Tasan, Gabriela Sustackova, Liguo Zhang, Jiah Kim, Mayandi Sivaguru, Mohammad HamediRad, Yuchuan Wang, Justin Genova, Jian Ma, Andrew S. Belmont, Huimin Zhao
A Cautionary Tail: Changes in Integrin Behavior with Labeling. Catherine G. Galbraith, Michael W. Davidson, James A. Galbraith
Printed Droplet Microfluidics for on demand dispensing of picoliter droplets and cells. Russell H Cole, Shi-Yang Tang, Christian A Siltanen, Payam Shahi, Jesse Q Zhang, Sean Poust, Zev J Gartner, Adam R Abate
Imaging action potential in single mammalian neurons by tracking the accompanying sub-nanometer mechanical motion. Yunze Yang, Xian-Wei Liu, Hui Yu, Yan Guan, Shaopeng Wang, Nongjian Tao
The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Christian Zierhut, Hironori Funabiki
Quantifying cell adhesion dynamics using a custom-made fluid shear device: experiments and model. Renu Maan, Gautam I Menon, Pramod A Pullarkat
Three-dimensional single cell patterning for mid- to high-throughput studies of tumor cell and extracellular matrix heterogeneity. Xiangyu Gong, Kristen L. Mills
Traction force screening enabled by compliant PDMS elastomers. Haruka Yoshie, Newsha Koushki, Rosa Kaviani, Kavitha Rajendran, Quynh Dang, Amjad Husain, Sean Yao, Chuck Li, John K. Sullivan, Magali Saint-Geniez, Ramaswamy Krishnan, Allen Ehrlicher
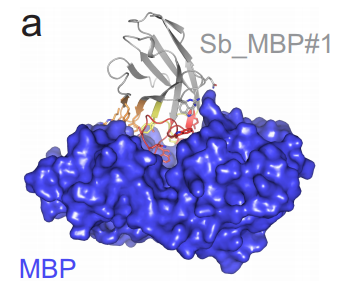
Synthetic single domain antibodies for the conformational trapping of membrane proteins. Iwan Zimmermann, Pascal Egloff, Cedric Hutter, Peter Stohler, Nicolas Bocquet, Melanie Hug, Martin Siegrist, Lisa Svacha, Jeniffer Gera, Samira Gmuer, Peter Spies, Daniel Gygax, Eric R. Geertsma, Roger J. P. Dawson, Markus A. Seeger
| Genome tools
A GAL80 collection to nullify transgenes in Drosophila olfactory sensory neurons. Jessica Eliason, Ali Afify, Christopher Potter, Ichiro Matsumura
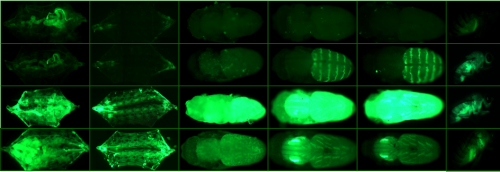
Regulating the UAS/GAL4 system in adult Drosophila with Tet-off GAL80 transgenes. Taylor Barwell, Brian DeVeale, Luc Poirier, Jie Zheng, Frederique Seroude, Laurent Seroude
Redesigning chromosomes to optimize conformation capture (Hi-C) assays. Heloise Muller, Vittore Scolari, Guillaume Mercy, Nicolas Agier, Stephane Descorps-Declere, Gilles Fischer, Julien Mozziconacci, Romain Koszul
Cell Type Specific Survey of Epigenetic Modifications by Tandem Chromatin Immunoprecipitation Sequencing. Mari Mito, Mitsutaka Kadota, Kaori Tanaka, Yasuhide Furuta, Kuniya Abe, Shintaro Iwasaki, Shinichi Nakagawa
Engineering post-translational proofreading to discriminate non-standard amino acids. Aditya M Kunjapur, Devon A Stork, Erkin Kuru, Oscar Vargas-Rodriguez, Matthieu Landon, Dieter Söll, George M Church
Linear Assembly of a Human Y Centromere using Nanopore Long Reads. Miten Jain, Hugh E. Olsen, Daniel J. Turner, David Stoddart, Kira V. Bulazel, Benedict Paten, David Haussler, Huntington Willard, Mark Akeson, Karen H. Miga
Resolving systematic errors in widely-used enhancer activity assays in human cells enables genome-wide functional enhancer characterization. Felix Muerdter, Łukasz M Boryń, Ashley R Woodfin, Christoph Neumayr, Martina Rath, Muhammad A Zabidi, Michaela Pagani, Vanja Haberle, Tomáš Kazmar, Rui R Catarino, Katharina Schernhuber, Cosmas D Arnold, Alexander Stark
DroNc-Seq: Deciphering cell types in human archived brain tissues by massively-parallel single nucleus RNA-seq. Naomi Habib, Anindita Basu, Inbal Avraham-Davidi, Tyler Burks, Sourav R Choudhury, Francois Aguet, Ellen Gelfand, Kristin Ardlie, David A Weitz, Orit Rozenblatt-Rosen, Feng Zhang, Aviv Regev
P53 toxicity is a hurdle to CRISPR/CAS9 screening and engineering in human pluripotent stem cells. Robert J. Ihry, Kathleen A. Worringer, Max R. Salick, Elizabeth Frias, Dan Ho, Kraig Theriault, Sravya Kommineni, Julie Chen, Marie Sondey, Chaoyang Ye, Ranjit Randhawa, Tripti Kulkarni, Zinger Yang, Gregory McAllister, Carsten Russ, John Reece-Hoyes, William Forrester, Gregory R. Hoffman, Ricardo Dolmetsch, Ajamete Kaykas
Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Ryan Marshall, Colin S. Maxwell, Scott P. Collins, Michelle L. Luo, Thomas Jacobsen, Chase Beisel, Vincent Noireaux
Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Alexandre Paix, Andrew Folkmann, Daniel H. Goldman, Heather Kulaga, Michael Grzelak, Dominique Rasoloson, Supriya Paidemarry, Rachel Green, Randall Reed, Geraldine Seydoux
Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Janice S. Chen, Yavuz S. Dagdas, Benjamin P. Kleinstiver, Moira M. Welch, Lucas B. Harrington, Samuel H. Sternberg, J. Keith Joung, Ahmet Yildiz, Jennifer A. Doudna
A CRISPR-Based Yeast Two-Hybrid System For Investigating RNA-Protein Interactions. Evan P. Hass, David C. Zappulla
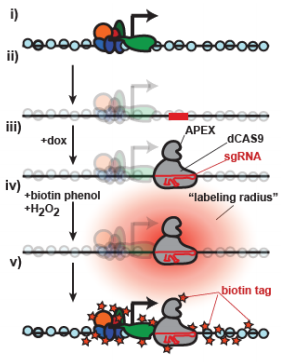
CRISPR/Cas9-APEX-mediated proximity labeling enables discovery of proteins associated with a predefined genomic locus in living cells. Samuel A. Myers, Jason Wright, Feng Zhang, Steven A. Carr
A reanalysis of Schaefer et al. does not indicate extensive CRISPR/Cas9 mediated off-target editing events. Reynald M. Lescarbeau, Bradley A. Murray, Thomas M. Barnes, Nessan Bermingham
“Unexpected mutations after CRISPR-Cas9 editing in vivo” are most likely pre-existing sequence variants and not nuclease-induced mutations. Caleb Lareau, Kendell Clement, Jonathan Y. Hsu, Vikram Pattanayak, J. Keith Joung, Martin J. Aryee, Luca Pinello
Response to Editas and Intellia: Unexpected mutations after CRISPR-Cas9 editing in vivo. Kellie A. Schaefer, Wen-Hsuan Wu, Benjamin W. Darbro, Diana F. Colgan, Stephen H. Tsang, Alexander G. Bassuk, Vinit B. Mahajan
Duplex Proximity Sequencing (Pro-Seq): A method to improve DNA sequencing accuracy without the cost of molecular barcoding redundancy. Joel Pel, Wendy W. Y. Choi, Amy O. Leung, Gosuke Shibahara, Laura Gelinas, Milenko Despotovic, W. Lloyd Ung, Andre Marziali
Fundamental limits on dynamic inference from single cell snapshots. Caleb Weinreb, Samuel Wolock, Betsabeh K. Tusi, Merav Socolovsky, Allon M. Klein
Mapping transcription factor occupancy using minimal numbers of cells in vitro and in vivo. Luca Tosti, James Ashmore, Boon Siang Nicholas Tan, Benedetta Carbone, Tapan K. Mistri, Valerie Wilson, Simon R. Tomlinson, Keisuke Kaji
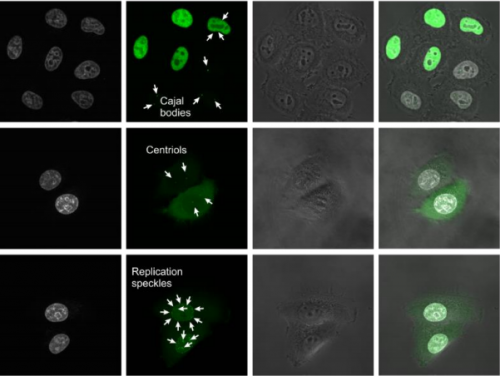
Versatile approach for functional analysis of human proteins and efficient stable cell line generation using FLP-mediated recombination system. Roman J. Szczesny, Katarzyna Kowalska, Kamila Klosowska-Kosicka, Aleksander Chlebowski, Ewelina P. Owczarek, Zbigniew Warkocki, Tomasz M. Kulinski, Dorota Adamska, Kamila Affek, Agata Jedroszkowiak, Anna V. Kotrys, Dominik Cysewski, Rafal Tomecki, Pawel S. Krawczyk, Lukasz S. Borowski, Andrzej Dziembowski
Quantifying homologous proteins and proteoforms. Dmitry Malioutov, Tianchi Chen, Jacob Jaffe, Edoardo Airoldi, Steven Carr, Bogdan Budnik, Nikolai Slavov
Protein identification with a nanopore and a binary alphabet. G Sampath
| Resources
Genome-wide genetic data on ~500,000 UK Biobank participants. Clare Bycroft, Colin Freeman, Desislava Petkova, Gavin Band, Lloyd T Elliott, Kevin Sharp, Allan Motyer, Damjan Vukcevic, Olivier Delaneau, Jared O’Connell, Adrian Cortes, Samantha Welsh, Gil McVean, Stephen Leslie, Peter Donnelly, Jonathan Marchini

ScreenCube: A 3D Printed System For Rapid and Cost-Effective Chemical Screening in Adult Zebrafish. Adrian T. Monstad-Rios, Claire J. Watson, Ronald Y. Kwon
OMICtools: a community-driven search engine for biological data analysis. Helene Perrin, Marion Denorme, Julien Grosjean, OMICtools community, Emeric Dynomant, Vincent J. Henry, Fabien Pichon, Stefan Darmoni, Arnaud Desfeux, Bruno J. Gonzalez
VolcanoR – web service to produce volcano plots and do basic enrichment analysis. Vladimir Naumov, Ivan Balashov, Vadim Lagutin, Pavel Borovikov, Alexey Alexeev
CellNetVis: a web tool for visualization of biological networks using force-directed layout constrained by cellular components. Henry Heberle, Marcelo Falsarella Carazzolle, Guilherme P. Telles, Gabriela Vaz Meirelles, Rosane Minghim
Science Game Lab: tool for the unification of biomedical games with a purpose. Benjamin Good, Sarah Santini, Margaret Wallace, Nicholas Fortugno, John Szeder, Patrick Mooney, Jerome Waldispuhl, Ginger Tsueng, Andrew I. Su
Use of a Sibling Subtraction Method (SSM) for Identifying Causal Mutations in C. elegans by Whole-Genome Sequencing. Braveen B. Joseph, Nicolas A Blouin, David S Fay
Objective, Quantitative, Data-Driven Assessment of Chemical Probes. Albert A. Antolin, Joe E. Tym, Angeliki Komianou, Ian Collins, Paul Workman, Bissan Al-Lazikani
A computational modelling framework to quantify the effects of passaging cell lines. Wang Jin, Catherine J. Penington, Scott W. McCue, Matthew J. Simpson
Effective normalization for copy number variation in Hi-C data. Nicolas Servant, Nelle Varoqaux, Edith Heard, Jean-Philippe Vert, Barillot Emmanuel
Hierarchical Analysis of Multi-mapping RNA-Seq Reads Improves the Accuracy of Allele-specific Expression. Narayanan Raghupathy, Kwangbom Choi, Matthew J Vincent, Glen L Beane, Keith Sheppard, Steven C Munger, Ron Korstanje, Fernando Pardo Manuel de Villena, Gary A Churchill
Accurate detection of complex structural variations using single molecule sequencing. Fritz J Sedlazeck, Philipp Rescheneder, Moritz Smolka, Han Fang, Maria Nattestad, Arndt von Haeseler, Michael Schatz
BGP: Branched Gaussian processes for identifying gene-specific branching dynamics in single cell data. Alexis Boukouvalas, James Hensman, Magnus Rattray
GEM: A manifold learning based framework for reconstructing spatial organizations of chromosomes. Guangxiang Zhu, Wenxuan Deng, Hailin Hu, Rui Ma, Sai Zhang, Jinglin Yang, Jian Peng, Tommy Kaplan, Jianyang Zeng
PHASIS: A computational suite for de novo discovery and characterization of phased, siRNA-generating loci and their miRNA triggers. Atul Kakrana, Pingchuan Li, Parth Patel, Reza Hammond, Deepti Anand, Sandra Mathioni, Blake Meyers
miRNAtools: advanced training using the miRNA web of knowledge. Francisco J. Enguita
Splatter: Simulation Of Single-Cell RNA Sequencing Data. Luke Zappia, Belinda Phipson,Alicia Oshlack
FaStore – a space-saving solution for raw sequencing data. Łukasz Roguski,Idoia Ochoa,Mikel Hernaez, Sebastian Deorowicz
HapCHAT: Adaptive haplotype assembly for efficiently leveraging high coverage in long reads. Stefano Beretta, Murray Patterson, Simone Zaccaria, Gianluca Della Vedova, Paola Bonizzoni
scmap – A tool for unsupervised projection of single cell RNA-seq data. Vladimir Yu. Kiselev, Martin Hemberg
RNA Transcriptome Mapping with GraphMap. Kresimir Krizanovic, Ivan Sovic, Ivan Krpelnik, Mile Sikic
GUIDES: sgRNA design for loss-of-function screens. Joshua A. Meier, Feng Zhang, Neville Sanjana
GROOLS: reactive graph reasoning for genome annotation through biological processes. Jonathan Mercier, Adrien Josso, Claudine Medigue, David Vallenet
SQUID: Transcriptomic Structural Variation Detection from RNA-seq. Cong Ma, Mingfu Shao, Carl Kingsford
HTSSIP: an R package for analysis of high throughput sequencing data from nucleic acid stable isotope probing (SIP) experiments. Nicholas D Youngblut, Samuel E Barnett, Daniel H Buckley
unitas: the universal tool for annotation of small RNAs. Daniel Gebert, Charlotte Hewel, David Rosenkranz
Euplotid: A Linux-based platform to physically edit the genome. Diego Borges, Richard Young
Atropos: specific, sensitive, and speedy trimming of sequencing reads. John P Didion, Marcel Martin, Francis S Collins
Research practice
Best Practice Data Life Cycle Approaches for the Life Sciences. Philippa C. Griffin, Jyoti Khadake, Kate S. LeMay, Suzanna E. Lewis, Sandra Orchard, Andrew Pask, Bernard Pope, Ute Roessner, Keith Russell, Torsten Seemann, Andrew Treloar, Sonika Tyagi, Jeffrey H. Christiansen, Saravanan Dayalan, Simon Gladman, Sandra B. Hangartner, Helen L. Hayden, William W. H. Ho, Gabriel Keeble-Gagnère, Pasi K. Korhonen, Peter Neish, Priscilla R. Prestes, Mark F. Richardson, Nathan S. Watson-Haigh,Kelly L. Wyres, Neil D. Young,Maria Victoria Schneider
The data and analysis underlying NIH’s decision to cap research support lacked rigor and transparency: a commentary. A Cecile JW Janssens, Gary W Miller, K Venkat Narayan
Sci-Hub provides access to nearly all scholarly literature. Daniel S Himmelstein, Ariel R Romero, Stephen R McLaughlin, Bastian Greshake Tzovaras, Casey S Greene
Text mining of 15 million full-text scientific articles. David Westergaard, Hans-Henrik Stærfeldt, Christian Tønsberg, Lars Juhl Jensen, Søren Brunak
Wikipedia as a gateway to biomedical research: The relative distribution and use of citations in the English Wikipedia. Lauren A Maggio, John Willinsky, Ryan Steinberg, Daniel Mietchen, Joe Wass, Ting Dong
Cross-linking BioThings APIs through JSON-LD to facilitate knowledge exploration. Jiwen Xin, Cyrus Afrasiabi, Sebastien Lelong, Julee Adesara, Ginger Tsueng, Andrew I Su, Chunlei Wu
Automatic OpenAPI to Bio.tools Conversion. Egon Willighagen, Jonathan Mélius
Designing an intuitive web application for drug discovery scientists. Nikiforos Karamanis, Denise Carvalho-Silva, Jennifer A. Cham, Luca Fumis, Samiul Hasan, David Hulcoop, Gautier Koscielny, Michael Maguire, William Newell, ChuangKee Ong, Eliseo Papa, Andrea Pierleoni, Miguel Pignatelli, Sangya Pundir, Francis Rowland, Jessica Vamathevan, Xavier Watkins, Jeffrey C. Barrett, Ian Dunham
Why not…
Love, not food, could have paved the path for dog domestication: A lesson from free-ranging dogs. Debottam Bhattacharjee, Shubhra Sau, Jayjit Das, Anindita Bhadra
The York Gospels: a one thousand year biological palimpsest. Matthew D. Teasdale, Sarah Fiddyment, Jiří Vnouček, Valeria Mattiangeli, Camilla Speller, Annelise Binois, Martin Carver, Catherine Dand, Timothy P. Newfield, Christopher C. Webb, Daniel G. Bradley, Matthew J. Collins
Primary souring: a novel bacteria-free method for sour beer production. Kara Osburn, Justin Amaral, Sara R. Metcalf, David M. Nickens, Cody M. Rogers, Christopher Sausen, Robert Caputo, Justin Miller, Hongde Li, Jason M. Tennessen, Matthew L. Bochman


 (No Ratings Yet)
(No Ratings Yet)