Living up to your cnidarian potential
Posted by Áine Varley, on 22 June 2023
In this post I will discuss our recent paper titled “Pluripotent, germ cell competent adult stem cells underlie cnidarian regenerative ability and clonal growth” as well as detailing the trials and tribulations that preceded it (Varley, et al., 2023).
Hydrozoan cnidarians have a population of stem cells, known as i-cells. The developmental potential of only one other hydrozoan i-cell has been described before this paper. Hydra‘s i-cells can give rise to the neuroglandular lineage and to germ cells. Their ectodermal and endodermal epithelial cells are self-renewing, constituting separate lineages. i-cells multipotent but not pluripotent (Bosch & David, 1987).
In terms of Hydractinia, it was established that a population of stem cells (known as i-cells) were able to give rise to every single lineage (Müller, et al., 2004). This showed that as a population, i-cells can give rise to the somatic and germ cell lineages. The potential of a single i-cell, however, was still unknown. Either i-cells had their own distinct lineage restricted sub-populations or they were pluripotent, and one i-cell could give rise to all cell types.
The aim of my research was to transplant a single i-cell and observe it in vivo. For this purpose, we had an extremely useful fluorescent reporter animal at our disposal. This animal had two fluorescent transgenes, the first transgene was Piwi1- driven GFP which is expressed in only i-cells and germ cells. It is downregulated in differentiated cells. The other transgene was Beta-tubulin- driven mScarlet. This transgene is expressed by all differentiated cells but not i-cells. This allowed us to identify i-cells by their GFP expression and lack of mScarlet expression and identify the i-cells progeny by their mScarlet expression and dim GFP fluorescence due to the half-life of the GFP. The aim of the project was to transplant a single GFP positive i-cell into a wild type recipient and identify the resulting progeny that would be identifiable by their mScarlet expression.
Bottlenecks on bottlenecks
Transplanting a single cell was my first objective when I started this project as a 1st year PhD student. I figured that it should take me 3-4 months maximum to get it sorted, optimised and then I could press on with the actual experiments and start generating some (hopefully decent) data. I began by looking at what single cell transplantations had been performed before in comparable models and hoped that it would be just as successful in Hydractinia.
Firstly, I tried incorporation of a cell via aggregation. This is something that was established in Hydra (Murphy & David, 1977). It required dissociation of full Hydractinia polyps. I could then identify a single i-cell by its GFP fluorescence and its lack of mScarlet fluorescence. This would then be picked up by a needle attached to a micro-injector and ejected into a tube full of dissociated wild type cells. These would then be centrifuged into an aggregate and cultured till, ideally, a polyp with a single transplanted i-cell would be established. Turns out, betting on a single cell surviving aspiration and/or centrifugation is a long shot. In meetings with my PI, Uri, I frustratedly referred to it as “a bottleneck on a bottleneck”.
After, countless amounts of failed transplantations via aggregation I was reinspired by the planarian community’s favourite method of single cell transplantation: direct injection into the animal (Wagner, et al., 2011). Perhaps by directly delivering a single cell I could finally cross off my first objective. This required a steady hand to ensure you didn’t overshoot the piercing of the epidermis and inject a cell into the gastric cavity. Unfortunately, after injecting into adult polyps and larvae from all angles I realised that this method wasn’t going to be feasible either. I was just not seeing any i-cell engraftment. It may have been a combination of mechanical issues on my part, maybe unhappy cells or also Hydractinia having a much more confined stem cell niche area than planarians do. Ultimately, we never found out why. We just knew it wasn’t a way forward to achieve our aims. At this point, I was almost 2 years into my PhD and that first objective was still not completed. The pressure was mounting, and I started considering alternative projects in the worst-case scenario. Whatever came next would be my last attempt at getting this project off the ground.
Keeping it in the niche
Finally, we had our own form of a eureka moment. We had been dissociating the animals to isolate one cell (like the planarian and Hydra communities had previously done) but we had overlooked something major and wonderful about Hydractinia. We had overlooked what sets them apart from both Hydra and planarians. Hydractinia is a colonial animal that can fuse and swap cells with a histocompatible relative via stolonal networks. I had been working to establish a method of transplanting cells when our model already had one itself. We could keep our i-cells within their own niche! I just had to find a way to effectively leverage it to my aims.
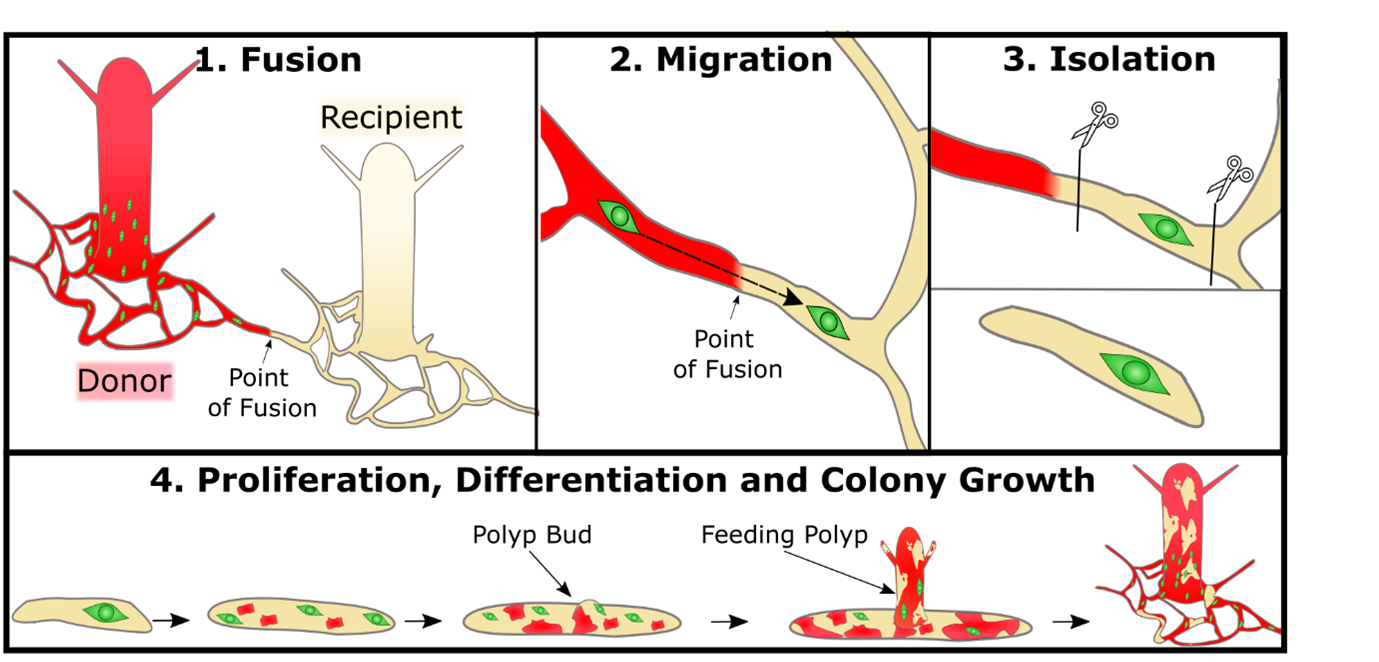
This began with me setting up Hydractinia “neighbourhoods”: a donor colony settled next to a recipient colony. The donor colony would grow and then fuse with the recipient by extended stolons and I could patiently observe i-cells migrating into the recipient tissue, waiting for a perfect moment to strike and isolate one cell (Figure 1). I had noticed that when the colonies were fed less, they were more likely to grow long branches of stolons rather than extending their mat. A long branch was preferable as it meant the i-cells could be transplanted “in single file” rather than like an encroaching army that was more unpredictable and difficult to follow a single cell. After numerous fusions of donor and recipient I isolated a single donor i-cell in a piece of recipient stolon tissue. The next day there were two i-cells. Then four. Then there were mScarlet expressing somatic cells AND i-cells. My transplanted cell was proliferating and differentiating. This new chimeric piece of stolon grew into a small colony with both wild type cells and cells derived from the one i-cell. Extraordinarily relieved, I checked off that first objective.
At the early stages there was a split between recipient and donor derived cells in the colony. To ensure that the donor cells were not outcompeted I performed “selective pruning”. This involved excising large portions of tissue that had only/mainly wild type recipient cells. This gave us a lot of peace of mind as we were more confident that the donor cells wouldn’t be outcompeted before they had developed sexual polyps.
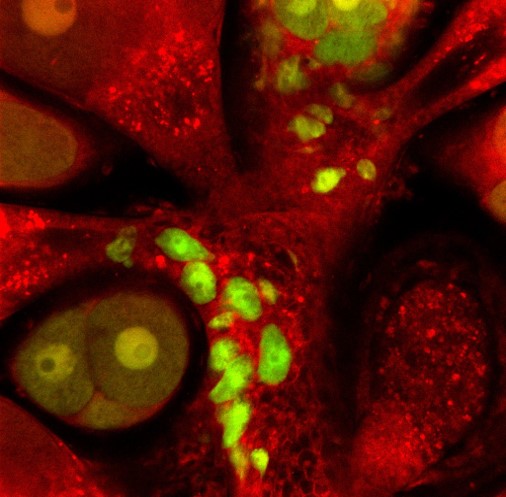
When the animal was sexually mature, we were excited to see sexual polyps that were male (from the recipient) but also fluorescent and female (from the donor). Some sexual polyps were even both. These donor derived GFP+ and mScarlet+ oocytes were able to be fertilised and gave rise to a new primary polyp that inherited the transgenes from the single donor i-cell (Figure 2). From this we knew that one i-cell can contribute to the next generation of Hydractinia.
Using live confocal microscopy on polyps cut from the colony, we were able to identify mScarlet+ neurons, epithelial cells, nematocytes and i-cells by their characteristic morphologies. Gland cells, however, were more difficult to identify. We didn’t have any antibodies that could identify them and even by morphology they often look like more vesicular epithelial cells. To address this, we had to look at it from a different perspective. At this later stage of the colony, we were able to confirm by flow cytometry that all cells present were transgenic and there were no recipient cells remaining. This meant that all cells, including gland cells, came from the single donor i-cell. This was the ultimate proof of pluripotency for us. There was not a single cell in the animal that one i-cell did not give rise to.
Don’t doubt your model
In humans, pluripotency occurs for a brief snippet of time pre-gastrulation. This paper established that Hydractinia stem cells are pluripotent for the full extent of their life. When starting, I didn’t expect for one single cell to be as globally influential as it was. In retrospect, I should have never doubted our model and its ability to impress.
References
Bosch, T. & David, C., 1987. Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Developmental Biology, pp. 182-191.
Müller, W. A., Teo, R. & Frank, U., 2004. Totipotent migratory stem cells in a hydroid. Developmental Biology, Volume 275, pp. 215-224.
Murphy, S. & David, C. N., 1977. Characterisation of interstitial stem cells in hydra by cloning. Developmental Biology, 58(2), pp. 372-383.
Varley, Á. et al., 2023. Pluripotent, germ cell competent adult stem cells underlie cnidarian regenerative ability and clonal growth. Current Biology, 33(10), pp. 1883-1892.
Wagner, D. E., Wang, I. E. & Reddien, P. W., 2011. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science, 332(6031), pp. 811-816.


 (1 votes)
(1 votes)