Making of the nerve cord: The story of telling neural progenitors when and where not to divide
Posted by Ignacio Monedero, on 5 May 2017
Comment on “Anterior-Posterior Gradient in Neural Stem and Daughter Cell Proliferation Governed by Spatial and Temporal Hox Control”, Current Biology 27, 1161-1172 (2017).
Ignacio Monedero, Behzad Yaghmaeian, Stefan Thor.
Department of Clinical and Experimental Medicine, Linkoping University, Sweden.
How cells are generated in proper numbers in order to form specific structures in an organism has been a subject of interest for many years. This issue is particularly intriguing in in the central nervous system (CNS), where the number of neurons of each type plays a crucial role in neural circuitry and cognitive capabilities in different species across evolution1. An intriguing feature of the CNS is that different regions have different cell numbers, and the number of neurons along the anterior-posterior (A-P) axis seems to be organized in a wedge-like appearance: the anterior part contains more neurons than the posterior part. This overall feature of CNS organization holds true for many vertebrate and invertebrate species. So how is the generation of proper number of neurons determined in the different regions of the central nervous system? This was, and still is, one of the questions around when I joined Stefan Thor’s lab in Linkoping, and the one which we try to shed light on in this post.
The total number of cells generated in a certain region is determined by several factors: the initial number of progenitors, the number of divisions that those progenitors go through, the proliferative behavior of the daughter cells produced by progenitors, and programmed cell death (PCD) present within the lineage. Drosophila CNS is subdivided into the brain, and the ventral nerve cord (VNC), equivalent to the mammalian and spinal cord respectively. Recent studies in the Drosophila VNC have provided an essential framework for addressing how the proper number of cells are generated in each segment. First, the recent meticulous mapping of the number of neural progenitors or neuroblasts (NBs) in each of the 13 segments of the VNC 2–6; second, our previous discovery of different daughter proliferation modes in NB lineages, where NBs initially bud off daughters that divide once, to generate two neurons/glia, denoted Type I, and subsequently switch to generating daughters that do not divide, instead directly differentiating, denoted Type 0 7; third, the identification of mutants that lack PCD in embryonic stages8. These findings gave us the opportunity to make a global and systematic study of proliferation in the VNC.
With this set up, our first challenge was to establish just how different the anterior segments are from the posterior ones, and that was one of the most exciting parts in the project. The 13 segments of the VNC are generated by a similar number of progenitors, about 64 per segment, in each segment. Our cell number analysis at the end of neurogenesis revealed that these 64 NBs generate different numbers of cells in different segments, with more cells in anterior segments than posterior ones. Based upon this finding we postulated two main processes that could contribute to this wedge-like appearance: removal of excess cells, by PCD, in posterior segments, and/or increased proliferation in anterior segments. Looking at PCD mutants, the A-P differences in numbers were still noticeable, suggesting PCD could not explain the difference. In contrast, we observed increased proliferation, of both NBs and daughters, in anterior segments, which pointed to proliferation as the main contributor to the A-P differences. To further address this notion, we analyzed proliferation in specific NB lineages, serially presented in all VNC segments. Whereas in anterior segments, NBs undergo more rounds of division and display a late or absent Type I->0 daughter proliferation switch, in the posterior segments NBs undergo fewer rounds of divisions and display an earlier Type I->0 switch. The combination of cell number analysis with global and single-lineage proliferation analysis allowed us to extract an average lineage behavior for thoracic segments versus posterior segments.
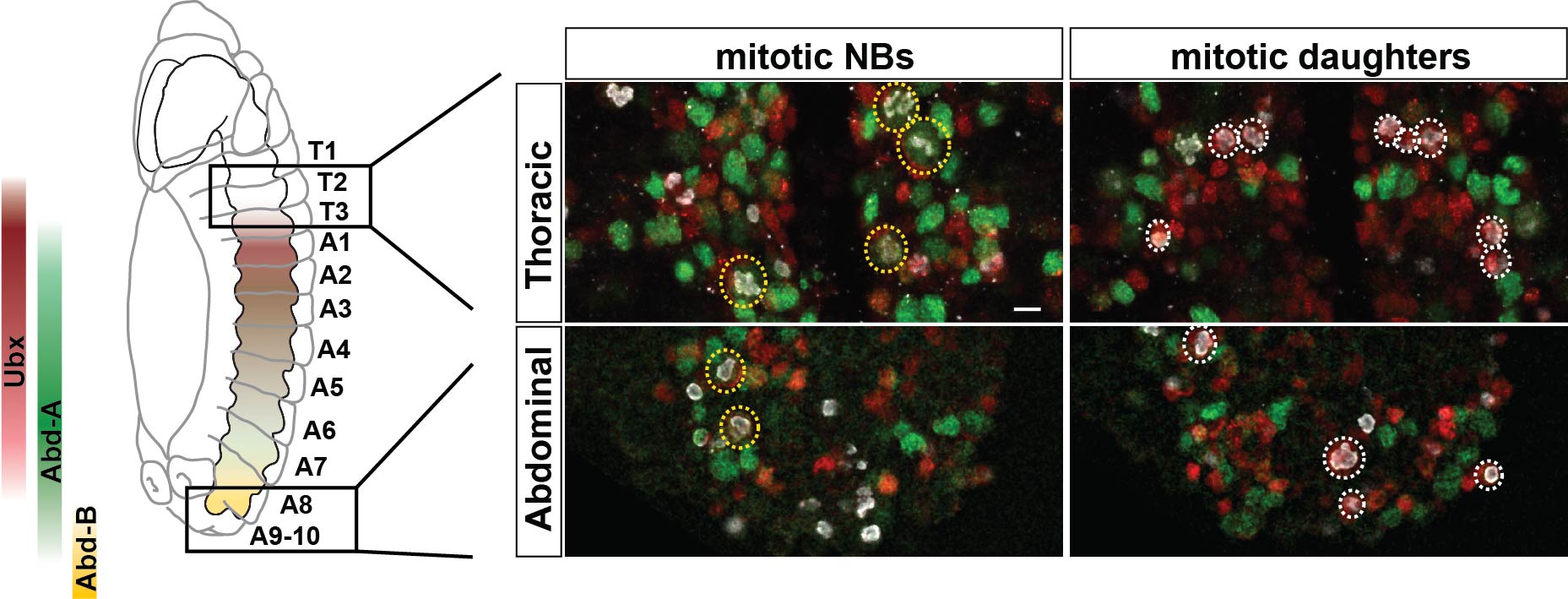
The next goal was to begin identifying some of the players controlling this gradient in proliferation behavior. The A-P axis differences obviously pointed to homeotic domains, and our previous work has identified the Hox gene Antennapedia (Antp) as one of the main triggers of the Type I->0 daughter proliferation switch in the thoracic segments7. Therefore, we analyzed the role of the more posteriorly acting Hox genes of the Bithorax-Complex (BX-C); Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B). Strikingly, we indeed observed that BX-C genes were important for the earlier Type I->0 daughter proliferation switch and NB proliferation exit observed in posterior segments. However, the triggering of the Type I->0 switch during later stages of lineage progression implied that the action of BX-C was gated in a temporal manner. Our previous studies of Antp had revealed that its expression in thoracic NBs commences during later stages. Studying this, we found that although Hox genes are indeed expressed in an A-P manner during early embryonic stages, early NBs display little if any BX-C protein expression. As NBs lineage progression proceeds, we observed onset of Hox protein expression, establishing the characteristic overlapping domains along the A-P axis, and this late onset in NBs then promotes the Type I->0 daughter proliferation switch. Based upon these findings we propose a model where at early stages NBs progress in a Hox-free “ground state”, allowing for high proliferative modes, whereas at later stages, the onset of Hox expression in NBs promotes the Type I->0 daughter proliferation switch, and eventually NBs stop dividing. This action of Hox genes establishes a gradient in NB lineage size, and results in the A-P wedge appearance.
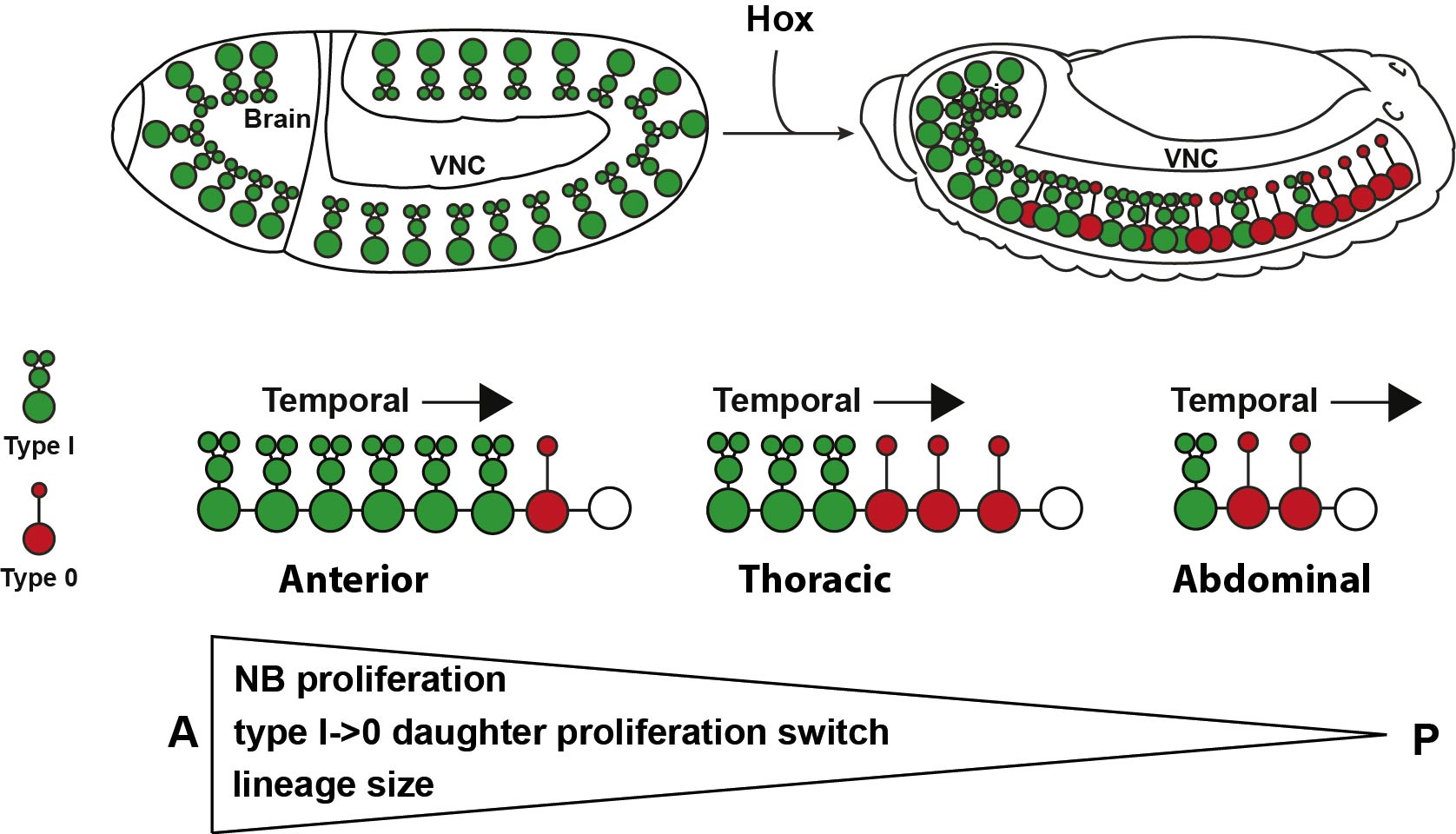
The hypothesis of the ground state is not new in Drosophila, and has been proposed by Fernando Casares and Richard Mann for appendages9. This idea proposes a “default” program, that we believe applies also to NB proliferation behavior, in thoracic segments, which is modified along the A-P axis by the same cues that determine the body plan. This program allows for homologous NBs in different segments to modify their proliferation behavior to generate the proper number of cells needed for that specific segment.
Since Hox genes are evolutionary conserved, establishing the body plan of invertebrates and vertebrates equally, it is reasonable to envision that they may have a conserved function in proliferation control in the nervous system. Indeed, several studies have identified Hoxc6, Hoxb9 and Hoxb13 related to suppression of proliferation in the vertebrate CNS10,11. Most intriguingly, while our current study was focused on the VNC, the most obvious wedge-like appearance in nervous systems is brain size when compared to nerve cord. Given that we have shown that Hox expression suppresses neural progenitor and daughter proliferation, and that the brain is a Hox free region, it seems reasonable to think that Hox genes could be one of the motors of the evolutionary expansion of the brain.
We are now very enthusiastic about the open possibilities of what is happening in the brain. The work is now focused on addressing how proliferation is controlled in the brain, and to what extent could alternate neural progenitor and daughter cell proliferation modes be a common feature, using similar or homologous tools, to establish A-P differences across different species.
- Herculano-Houzel, S. Not All Brains Are Made the Same: New Views on Brain Scaling in Evolution. Brain. Behav. Evol.78, 22–36 (2011).
- Birkholz, O., Rickert, C., Berger, C., Urbach, R. &Technau, G. M. Neuroblast pattern and identity in the Drosophila tail region and role of doublesex in the survival of sex-specific precursors. Development 140, 1830–1842 (2013).
- Schmid, A., Chiba, A. & Doe, C. Q. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126, 4653–4689 (1999).
- Schmidt, H. et al. The Embryonic Central Nervous System Lineages ofDrosophila melanogaster. Dev. Biol. 189, 186–204 (1997).
- Bossing, T., Udolph, G., Doe, C. Q. &Technau, G. M. The Embryonic Central Nervous System Lineages ofDrosophila melanogaster. Dev. Biol. 179, 41–64 (1996).
- Wheeler, S. R., Stagg, S. B. & Crews, S. T. MidExDB: A database of DrosophilaCNS midline cell gene expression. BMC Dev. Biol. 9, 56 (2009).
- Baumgardt, M. et al. Global Programmed Switch in Neural Daughter Cell Proliferation Mode Triggered by a Temporal Gene Cascade. Dev. Cell 30, 192–208 (2014).
- Tan, Y. et al. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development 138, 2197–2206 (2011).
- Casares, F. & Mann, R. S. The Ground State of the Ventral Appendage in Drosophila. Science 293, 1477–1480 (2001).
- Epstein, M., Pillemer, G., Yelin, R., Yisraeli, J. K. &Fainsod, A. Patterning of the embryo along the anterior-posterior axis: the role of the caudal genes. Development 124, 3805–3814 (1997).
- Economides, K. D., Zeltser, L. &Capecchi, M. R. Hoxb13 mutations cause overgrowth of caudal spinal cordand tail vertebrae. Dev. Biol. 256, 317–330 (2003).


 (1 votes)
(1 votes)