Of Arms and Legs: Shedding light onto developmental gene regulation
Posted by BKragesteen, on 19 November 2018
Written and illustrated by: Bjørt K. Kragesteen, Malte Spielmann, and Guillaume Andrey.
In early development, the forelimb and hindlimb buds of tetrapods are morphologically uniform. However, as limb development proceeds, each individual tissue attains a characteristic morphology that ultimately defines the identity of a forelimb (arm) or a hindlimb (leg). How do undifferentiated limbs bend their morphogenetic trajectories into arm and legs, since they are patterned by similar developmental processes? It is commonly accepted that the differential activities of a handful of genes instruct the formation of either arms or legs; yet the mechanism leading to their differential regulation in fore- and hindlimb buds was unknown. In our recent study, we dissected in detail such regulatory mechanisms and have revealed, once again, how important the function of the non-coding regulatory genome is, representing the overwhelming majority of vertebrate genomes.
In Stefan Mundlos’ research group at the Max Planck Institute for Molecular Genetics in Berlin, the focus is on the lessons that human limb malformations can teach us about gene regulation. Most research projects start with detecting the genomic mutation in human patients with congenital limb malformations at the Charité Berlin university hospital. These limb malformations are frequently caused by large changes in the DNA called structural variants including: deletions (removing information), inversions (displacing information), and duplications (doubling and misplacing information). One can imagine that such mutations might mess up the genetic information, and by detecting them in patient genomes, structural variants can provide a clue as to where important non-coding regulatory elements are located. From this starting point, it is possible to look for neighbouring genes that become misregulated following the mutation, ultimately changing the developmental program and the formation of the embryo.
Our story started back in 2010 when Malte Spielmann, a clinical scientist interested in structural variants and congenital disease, tried to work out the genetic cause in three families with a very rare malformation syndrome affecting the arms of the patients, known as Liebenberg syndrome (named after the doctor that first described it back in 1973). When he looked at the X-rays of the patients upside down, it struck him that the arms looked very much like legs! If we think about our arms and legs, they actually share many similarities: both are formed by one long bone (upper arm/thigh: the humerus/femur), followed by two bones (lower arm/leg: ulna/fibula and radius/tibia), terminating in many small bones (hands/feet: carpals, metacarpals, and phalanges) (see Figure 1).
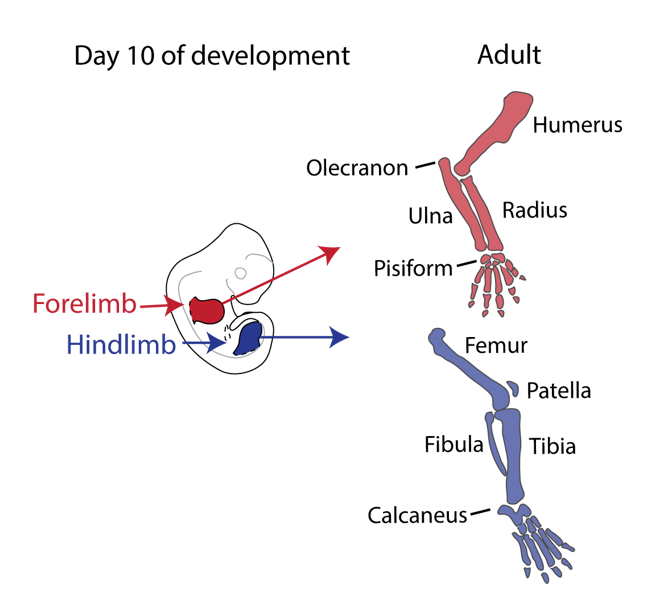
Nevertheless, they flex in opposite directions, as the arms have a specialised elbow joint, where the tip of the ulna called the olecranon (the pointy elbow) grabs around the distal end of the humerus. The legs have a specialised knee joint with a small bone, the patella (knee cap), ensuring stability and enabling weight bearing. However, the Liebenberg syndrome 3D CT scan showed the following: the fingers were very short (brachydactyly) looking very much like toes; the small bones of the wrist (pisiform, triquetral) were fused together looking like the heel bone (calcaneus), and the olecranon was reduced and no longer grabbing around the end of the humerus. Instead, the end of the humerus was broader as a patella like bone appeared fused to it (Figure 2).
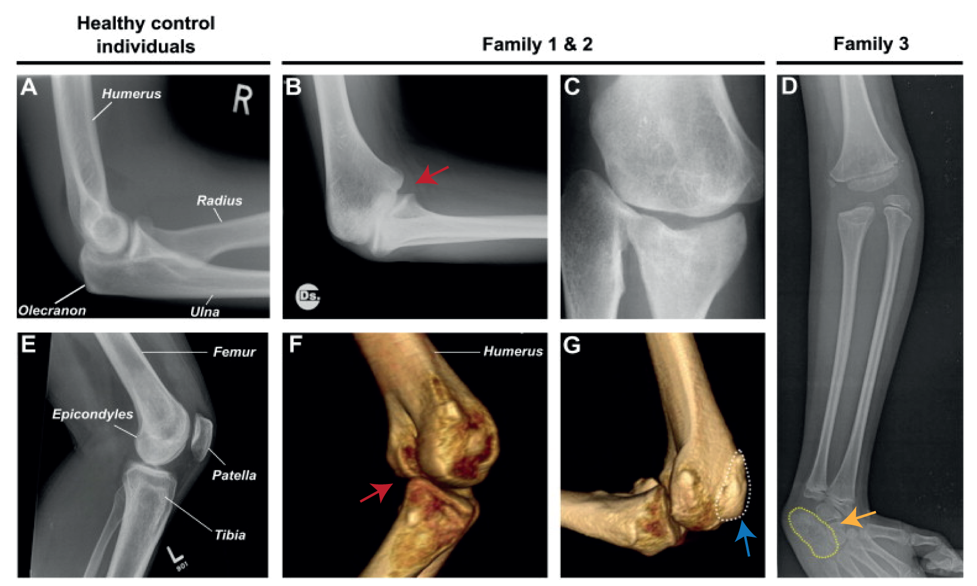
How on earth could this happen? Malte performed DNA analysis (array-CGH) and found a large deletion (107 kb) on chromosome 5 in all affected individuals. The deletions remove the gene H2AFY. However, this gene is a housekeeping gene and in mice in which the gene had been inactivated do not show any limb phenotype. Malte then looked on each side of the gene and found two interesting things:
On the centromeric side lies a gene named PITX1, which is expressed exclusively in the hindlimb during limb development and is known to be the only transcription factor that patterns the tissue. Inactivation of Pitx1 in mice results in reduction of leg morphology, such as loss of the patella and reduction of the calcaneus and the knee joint changing into an elbow-like joint. On the contrary, misexpression of Pitx1 in mouse forelimbs partially transforms the elbow joint into a knee like joint. Thus, Pitx1 misregulation in the forelimb seemed like a good candidate to explain to Liebenberg phenotype. However, Pitx1 regulation in tetrapods was unknown. Why would a deletion 200 kb upstream of the PITX1 promoter cause Liebenberg syndrome?
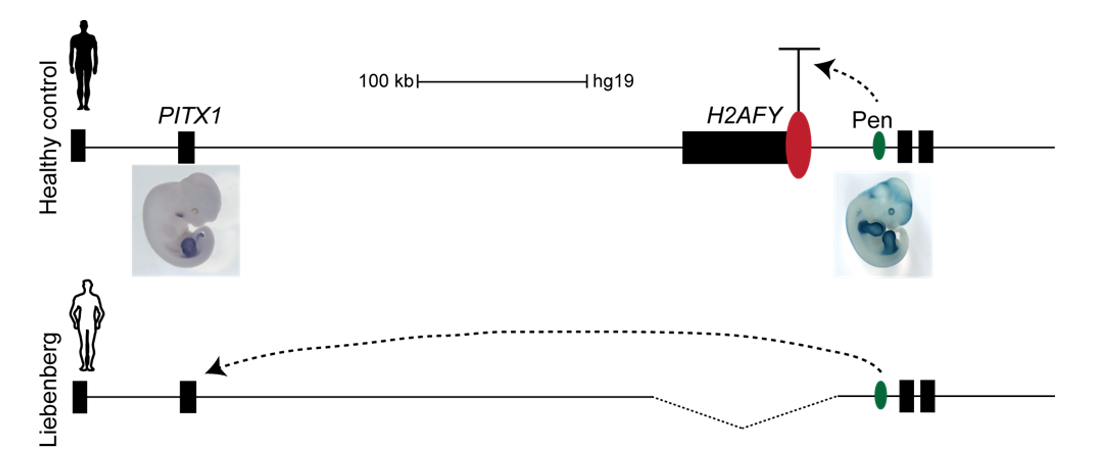
Interestingly, on the telomeric side of the deletion, 300 kb upstream of PITX1, a non-coding enhancer element can be found, called Pen (pan limb enhancer) that is active in both forelimbs and hindlimbs. Enhancer elements are defined as sequence specific stretches of DNA that are bound by transcription factors that dictate the activity and ensure communication with the correct gene promoters through chromatin folding. Often several enhancer elements regulate a target promoter and the collective activity of the tissue specific enhancer reflects the promoter transcriptional output. Recent development of proximity-ligation chromatin conformation capture technology (e.g. genome wide Hi-C) has demonstrated that the genome is partitioned into topological associating domains (TADs). TADs are scaffolds of preferential interactions between cognate promoters and enhancers and thus protect them from promiscuous activity from neighbouring TADs by boundary elements. These TADs were said to be stable across cell types and evolutionary conserved. With this in mind, the following hypothesis was formulated:
The Liebenberg deletion removes a TAD boundary element that normally separates PITX1 and Pen, resulting in PITX1 adopting a foreign enhancer, i.e. Pen, that is active in both fore- and hindlimbs. This enhancer adoption thus results in abnormal activation of PITX1 in the forelimb, where it should never be expressed, altering the patterning of the forelimb into a hindlimb, and partially transforming the arms into legs (Figure 3).
To test the hypothesis, Bjørt Kragesteen, a PhD student in the lab interested in deciphering non-coding functionality, sought to discern the molecular pathomechanism of Liebenberg syndrome together with Malte. First Bjørt created mouse mutants using CRISPR-Cas9 engineering that was newly established in the lab (back in 2013). She used two gRNAs to generate Liebenberg-like deletions and inversions at the mouse Pitx1 locus. Excitingly, analysis of Pitx1 mRNA expression in the early mutant mouse embryos showed Pitx1 misexpression in forelimbs! Skeletal analysis of adult mice showed a Liebenberg-like phenotype where the olecranon was reduced, an ectopic bone (patella-like) appeared at the humerus and the rotation of the arm was similar to the legs (Figure 4). Amazing. Case solved.
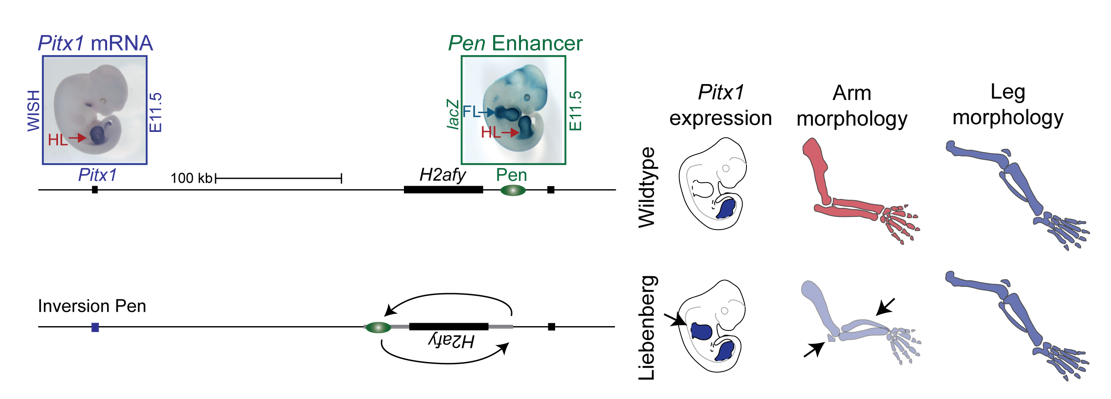
However, many questions remained unanswered. Why is Pen, a strong fore-and hindlimb enhancer, located relatively close to Pitx1? Isn’t that too risky for a key hindlimb patterning gene?
In parallel to Bjørt’s project, Guillaume Andrey, who started his postdoc in the laboratory in early 2014, was investigating the normal regulation of Pitx1, by combining deletions and inversions of its putative regulatory landscape and enhancer assays. He observed that several engineered structural variants, which did not alter the pre-supposed boundary between Pitx1 and Pen, resulted in a mirror image pattern between fore- and hindlimb, identical to the one of the Pen activity and a Liebenberg phenotype. This observation suggested that Pitx1 could be controlled by the Pen element even when the “boundary” was intact. From that point, it became evident that in hindlimbs Pen could play a role in the regulation of Pitx1.
We thus next deleted the Pen enhancer to see what would happen. A visit to the animal facility some weeks later revealed that some of the adult homozygous Pen deletion mice developed club feet, dragging their hindlimbs behind. Both mouse and human patients haploinsufficient for Pitx1 develop clubfeet. Moreover, the mutants showed reduced Pitx1 expression and skeletal abnormalities whereby the patella was missing (Figure 5). This was solid evidence that Pen indeed is a Pitx1 enhancer!
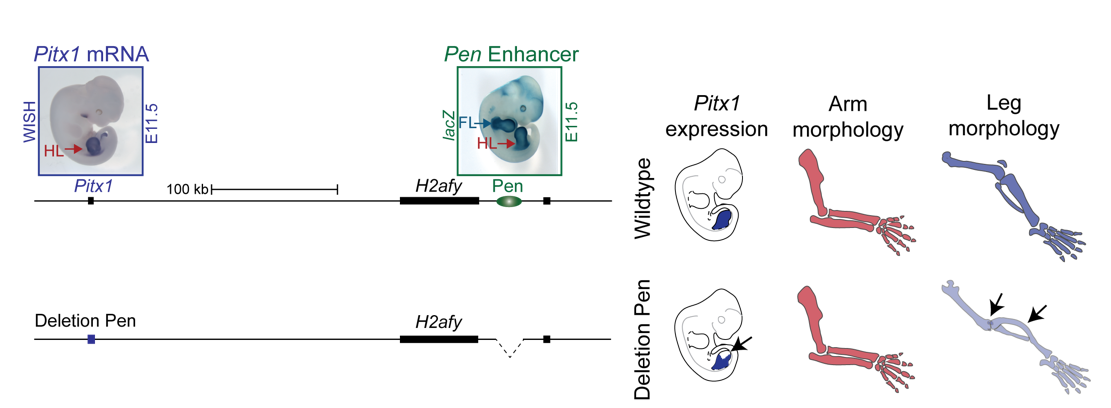
With this exciting finding we identified something not hereto described: A gene can be regulated by an unspecific long range enhancer; however, changing its location in the 2D regulatory landscape results in misregulation of the target gene and can transform tissue morphologies. We continued the search for a hindlimb specific Pitx1 enhancer, but no matter how much cloning and testing of enhancer elements we did, no hindlimb specific enhancer could be found. We thus decided to join forces, merging the projects about the normal and Liebenberg regulation of Pitx1, in order to understand how a gene that is solely expressed in the hindlimb can be controlled by unspecific enhancer elements active in both forelimbs and hindlimbs.
The research question thus became: what prevents Pitx1 expression in forelimbs in wildtype animals if its enhancer is active in both pairs of limbs?
The next obvious layer of gene regulation to scrutinise was the chromatin folding at the Pitx1 locus in limbs. We first employed circular chromatin conformation capture (4C) technology in forelimbs vs hindlimbs to detect which regions surrounding Pitx1 were in close proximity to the gene. But, using the Pitx1 promoter region as a viewpoint, no differences were observed. However, Guillaume was using a slightly alternative 4C viewpoint in the gene body, and using the same library could see very strong differences between the tissues and clear Pitx1-Pen interactions in hindlimbs, indicating that the locus structure was highly defined. This was further confirmed using capture-C variation of the method using a larger fragment as a “bait” to see the interactions with the Pitx1 promoter region. Excitingly, differences between forelimbs vs hindlimbs emerged whereby Pitx1 showed differential interactions with Pen: in hindlimbs, Pen and Pitx1 come into close proximity enabling its tissue specific activation, while in forelimbs they are kept separated! Yet, this did not give the complete picture. At that point in time (2016) a new C-method, capture Hi-C, was developed and we established it in the lab. Here RNA probes are used to pull down a region of interest, and we enriched 3 megabases surrounding the Pitx1 locus. This provides a more complete picture of the interactions over the whole locus. Contrasting forelimbs and hindlimbs interaction heat-maps showed a clear difference: in hindlimbs Pitx1 forms loops with several regions (RA1, RA3 and Pen), which were almost completely diminished in forelimbs, but with a forelimb specific loop with the repressed gene Neurog1 occurs.
Still, this type of analysis gave us a 2D image of what was going on at the locus and we had a hard time imagining what this looks like in 3D, despite many hand drawn depictions. We thus initiated a collaboration with physicists in Italy in Mario Nicodemi’s research group. They used our capture Hi-C data and ran computational simulations using a so-called strings and binders model. They sent us the modelling results and 3D videos of forelimbs vs hindlimbs as well as an inversion mutant that shows the Liebenberg phenotype. The result was jaw-dropping: in wildtype forelimbs, the locus forms an inactive conformation whereby Pitx1 and Pen are kept a part, and where the repressed Pitx1 is embedded within its own domain next with repressed Neurog1 (neither of the genes are active in forelimbs and thus are covered with repressive epigenetic marks and hang out together). In wildtype hindlimbs, the locus folds in three domains and now Pitx1 is sitting at the surface of its domain directly facing Pen, thus enabling communication and robust transcription. Finally, in mutant forelimbs, bringing Pen closer to Pitx1 results in active folding of the whole locus, such as that in hindlimbs. Thus, not only do Pen and Pitx1 interact ectopically in mutant forelimbs, but the 3D-conformation becomes hindlimb-like, resulting in ectopic Pitx1 expression and arm-to-leg transformation.
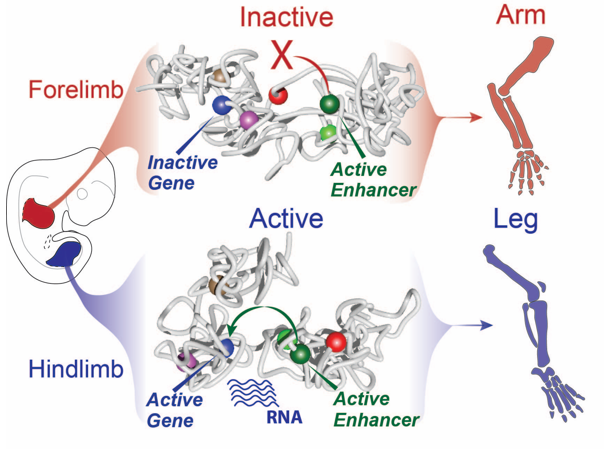
With all these data in hand and the genetic evidence that Pen was responsible when misplaced in the nuclear 3D space for the ectopic expression of Pitx1 in the “Liebenberg” forelimb, we could determine that the Pitx1 locus dynamic structure modulates the Pen activity in normal embryos. Specifically, in forelimb it represses the enhancer by keeping it away from Pitx1 promoter and that upon activation in hindlimb, it can participate in Pitx1 robust expression. Finally, it also showed that ectopic interaction between a gene and its own enhancer, following structural variant, in a process called gene endo-activation can cause gene misexpression and disease.
Read the full story:
Kragesteen B.K.*, Spielmann M.*, Paliou C., Heinrich V., Schöpflin R., Esposito A., Annunziatella C., Bianco S., Chiariello A.M., Jerković I., Harabula I., Guckelberger P., Pechstein M., Wittler L., Chan W.L., Franke M., Lupiáñez D.G., Kraft K., Timmermann B., Vingron M., Visel A., Nicodemi M., Mundlos S. and Andrey G.
Dynamic 3D Chromatin Architecture Contributes to Enhancer Specificity and Limb Morphogenesis.
Nat Genet. 2018 Doi: 10.1038/s41588-018-0221-x
*These authors contributed equally


 (No Ratings Yet)
(No Ratings Yet)