The people behind the papers: Miguel Brun-Usan & Isaac Salazar-Ciudad
Posted by the Node Interviews, on 4 January 2017
The tenth paper featured in this series comes from the first issue of Development for 2017, and uses computational modelling to investigate the importance of different cellular processes in spiral cleavage of the early embryo.
We caught up with the paper’s first author Miguel Brun-Usan and Isaac Salazar-Ciudad, group leader at the University of Helsinki in Finland, to find out more about the history of the work and how computational modelling can help developmental biologists.

So Isaac, can you give me your scientific biography and the main questions your lab is trying to address?
ISC I studied Biology in the Autonomous University of Barcelona. Some months before finishing, I started to look for research group in evolution in Spain. I found Pere Alberch‘s research and I immediately got hookeded by his approach. He approached evolution not from the reductionistic population genetics approach (that I had studied in detail), but from the complementary approach of evolution and development. I met him and he suggested me to contact Ricard V. Solé (at the time at the Polytechnic University of Catalonia) and Jordi Garcia Fernandez (at the University of Barcelona) since he was at the time already quite ill. I started my PhD with Ricard and Jordi on mathematical models of gene networks in evolution and development. During that time I also started to collaborate with Stuart A. Newman and Jukka Jernvall.
After my PhD, I went to Helsinki University to work as a Marie-Curie postdoctoral fellow with Jukka Jernvall in tooth morphogenesis and evolution. In 2008 I moved to the Autonomous University of Barcelona as a Ramón y Cajal fellow. This position was supposed to be tenure-track but with the financial crisis in Spain it soon became clear that it would not be tenured. In 2011 I moved back to the University of Helsinki as a junior group leader and Finnish Academy Fellow.
“The questions we aim to address in my lab are how the way development works affects the direction of evolutionary change and how development itself evolves”
The questions we aim to address in my lab are how the way development works affects the direction of evolutionary change and how development itself evolves. Development determines which kind of morphological variation is possible due to genetic mutation, and then it has a strong influence, together with natural selection, on how the morphology is going to change in evolution. To study that we build computational models of how genes and cells interact in networks to construct the body. We do that for specific organs in close collaboration with experimental groups (we have been working with mammalian teeth, fly wings, fly segmentation and turtle carapace) but also in general. We also set those models to evolve in the context of populations under natural selection to acquire general principles about how gene networks and development as a whole evolves.
What is science and life like in Helsinki?
ISC It is great. The funding situation has been, until the last elections, rather good (especially compared to Spain). Helsinki has in proportion to its small size a rather large and active scientific life with many high quality researchers. There is a relatively high concentration of researchers working in evo-devo and development (again in proportion to its size) and a long tradition in developmental biology and in evolutionary biology.
Helsinki has a very high quality of life according to many non-Finnish magazines (Newsweek, Daily Telegraph, etc…). I am originally from Barcelona that, like many South and Central European cities, is rather dense and devoid of green areas. Barcelona is good for visiting but not so good for living. Helsinki is the contrary. Living in Helsinki is like living in a large nice park. Even in the poorest neighbourhoods, one is always a few steps away from a forest, a park or the sea. In spite of its parkland nature, public transportation is quite good. In summer the city centre has a very active cultural live with many free outdoor concerts and large crowds of cheerful people relaxing in the parks. Winter is great if you like winter sports and snow, as I do. In addition, almost everybody speaks English so it is quite easy to have a normal life in spite of being a foreigner.
And Miguel, how did you come to work with Isaac?
MBU Five years ago, I finished a Master’s Degree in Evolutionary Biology in the Complutense University of Madrid, which gave me a grasp of why evo-devo is relevant to understand evolution. As a person who has been fascinated by biological evolution since childhood, I quickly started to look for a PhD position in a research group working on morphological evolution. Unfortunately, these groups are very scarce in Spain.
However, through an active search, I found in a Spanish webpage for graduated students (RedIris) an advert from Isaac which seemed to fulfil perfectly my academic interests. I had read some of his papers, but I did not know him personally, so I immediately contacted him and we had several interviews. After these interviews, Isaac agreed I had the right motivation and background to work in his group. I had no previous experience in programming.
Your paper describes computational models of cleavage. How can this sort of modelling complement descriptive and experimental embryology?
ISC & MBU The main way in which models can help descriptive and experimental embryology is by discarding existing hypotheses. In our case for example, the model allows to prove that some existing hypotheses can not lead to the cleavage patterns we observe, it is logically impossible. On the other hand models allow to suggest which hypotheses, alone or in combination, could in fact produce the observed patterns. Models, however, do not show that these suggested hypotheses are right. This latter thing always requires, ultimately, experiments but the model suggests which hypotheses, and then which experiments, are the most promising.
“The main way in which models can help descriptive and experimental embryology is by discarding existing hypotheses”
Do you think modelling is currently well-used and appreciated in the field?
ISC & MBU No. For the last ten years there has been a steady increase in the interest, and in the number of articles, in modelling in developmental biology. This is certainly good news but the advantages of modelling are not yet widely recognized and there is a widespread misunderstanding and misuse of modelling. In our view there are several reasons for it.
First, a large proportion of developmental biology is still mostly concerned with genes and not so much with processes. Genes are crucially involved in most developmental processes but are not, on their own, enough to explain any of them. Hypotheses about how a system develops are only possible when considering how genes interact, how cells interact or both. Then for most systems we do not know enough to even start to build a reasonable hypothesis, and so we can not build any models either. In many developmental systems we know a lot, typically a long list of genes important for it, but we are missing some crucial data, like which cell behaviours are used (cell division, cell adhesion, apoptosis, etc…), that is necessary to make predictive models.
Second, the difficulty understanding what models are for is usually not due to difficulties understanding models or mathematics as such, but to difficulties understanding hypothesis-driven research. Most research in developmental biology is, in practice, not hypotheses-driven. A large proportion of the research is devoted to identify interactions (usually genetic) that are necessary for the development of one or several organs. This exploratory work is certainly necessary but there should also be work (and there is in fact some) proposing hypotheses about how the identified interactions get coordinated to explain how a system develops, and proposing how to test these hypotheses. Models are helpful in these latter two things. Each model is simply a mathematical implementation of an hypothesis but, because of being mathematical, such implementation provides accurate predictions of how developmental variables of interest (for example organ shape) change over developmental time. These accurate predictions can then be compared with the observed development to falsify, or not, the hypotheses on which the model are built.
“For the last ten years there has been a steady increase in the interest in modelling in developmental biology. This is certainly good news but the advantages of modelling are not yet widely recognized and there is a widespread misunderstanding and misuse of modelling”
By exploring which hypotheses are consistent with experiments and which ones are not one learns. Models are not absolutely necessary for this hypothesis-driven research but they are quite useful for it and if one is not used to this hypothesis-driven research it is not possible to appreciate and use models. This relates to a common criticism to models: that models usually do not include all available data, or that their results are not totally realistic. This may be more the case for some models than for others, but the fact is that no model is perfect simply because no hypothesis is. It is only by making hypotheses and falsifying them that one can learn.
Third, many developmental biologists feel uneasy with programming or mathematics. Although the actual mathematics involved are often relatively simple (I for example taught myself the necessary maths for the models I do) this has created a niche for non-biologists to work in modelling in developmental biology or biology in general. From my experience as a reviewer that is often quite sub-optimal. This is specially the case for computer scientists and engineers since they are usually even less familiar with hypotheses-driven research and, in addition, their lack of a deep understanding of biology precludes them from evaluating which questions are relevant for biological theory and which ones are not. This adds to the clichés about the difficulty in communication between the people making the models and the experiments. I have avoided those by being myself a biologist and working only with experimentalists doing hypothesis-driven research.
What led you to focus on spiral cleavage in particular?
ISC & MBU Miguel was quite fascinated by the study of theoretical morphospaces as are often done in palaeontology. In these studies one can reproduce all the morphologies in a clade, for example the shape of the shell as in Raup’s classic article, by giving specific values to an equation. This equation is, however, merely phenomenological; it does not include any understanding of the processes by which those morphologies are built. Miguel wanted to do something similar but based on the rules of development, as Isaac had done in the past on a more restricted scale. The aim was then to be able to pinpoint which aspects of development have to change to reproduce the variation between many species. Miguel had a long lasting interest in very early development and we realized that, in fact, the developmental stage for which there is information about more species is cleavage. Originally Miguel wanted to do it for all kinds of cleavage but we set first for spiral cleavage because it is the most common cleavage type at the phylum level.
Can you give us the key results of your paper in a paragraph?
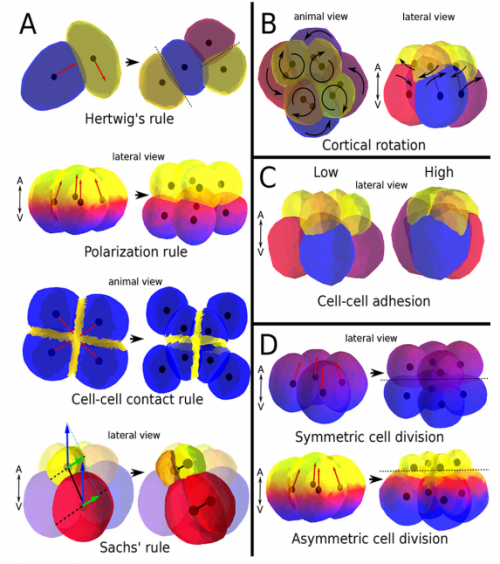
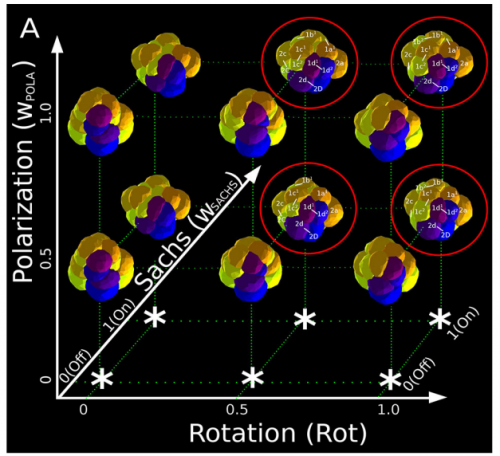
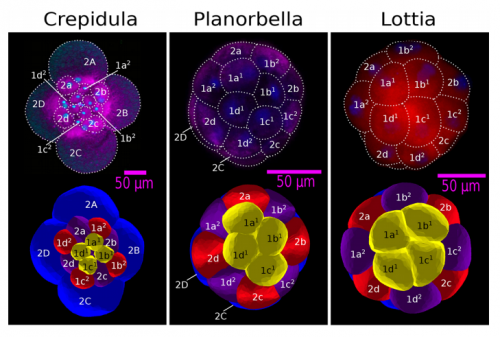
ISC & MBU Different cellular processes have been hypothesized to be responsible for the development of the specific spatial arrangement of blastomeres in the spiral blastula. These include the orientation of cell division according to an animal-vegetal gradient, according to cell’s main axis (Hertwig’s rule), according to the contact areas between cells or orthogonally to previous divisions (Sach’s rule). We use a computational model of cell and tissue bio-mechanics to implement the different existing hypotheses about how the specific spatial arrangement of cells in spiral cleavage arises during development. We found that cell polarization by an animal-vegetal gradient, a bias to perpendicularity between consecutive cell divisions (Sachs’ rule), cortical rotation and cell adhesion, when combined, reproduce the spiral cleavage while other combinations of processes can not. Specifically, cortical rotation is necessary in the 8-cell stage to displace all micromeres into the same direction, being this displacement random in direction if only cell adhesion is included. By varying the relative strength of these processes we reproduce the spatial arrangement of cells in the blastulae of seven different species (four snails, two polychaetes and a nemertean).
And what does your model suggest about the relationship between intercellular communication and spatial arrangement?
ISC & MBU One of the article’s results is that, in fact, the spiral cleavage, at least in its most generic form and also for the species we consider, does not require of any inductive events between cells (no diffusion of molecular factors between cells required). In other words, the relative spatial arrangement of cells does not require from cell communication. This does not mean that this communication does occur, we know it occurs in many species and this communication may be required to specify cell fate and, perhaps, for spiralian cleavages that depart significantly from the generic one.
Can your work tell us something about the evolution of cleavage patterns in animals?
ISC & MBU Bibliography shows us that most animal clades use the same set of basic (and evolutionarily old) cell processes for building up their cleavage patterns. Thus, even though our work is only concerned with spiral cleavage, our results can shed light in other, non-spiralian cleavage patterns. First, non-spiralian patterns have to be generated by other spatio-temporal combinations of similar cell processes, and variation within these non-spiralian patterns are very likely produced by variation in the strength of these other rules. Second, as is the case for the spiral pattern, some patterns may be more likely to arise from development than others (they require less cell processes, and/or less coordination between them). This may account for the uneven distribution of cleavage patterns among animals.
“Most animal clades use the same set of basic (and evolutionarily old) cell processes for building up their cleavage patterns. Thus, even though our work is only concerned with spiral cleavage, our results can shed light in other, non-spiralian cleavage patterns”
As someone who used to work in a ‘wet’ lab, I’m wondering about the process of modelling – how long does it take to run, and when do you know that you have an interesting result? Do you get ‘eureka!’ moments?
MBU I would like to make here a clear distinction between the model (SpiralMaker) itself and the in silico experiments we have performed with this model.
The model (or more precisely, its more general version EmbryoMaker), comprises more than ten thousand lines of programming language, organized in different inter-dependent modules. Most of it is not used in this article however. Such a big code has been built by different people working in Barcelona and in Helsinki during four years (specifically, I programmed many parts of the code relevant for non-epithelial cells). The most challenging aspect of this stage was to implement the different biological phenomena in a mathematical model and to make the model work as a coherent whole. Finally, we had to check that each implemented biological phenomenon worked realistically, but there was little room for Eureka! moments at this stage.
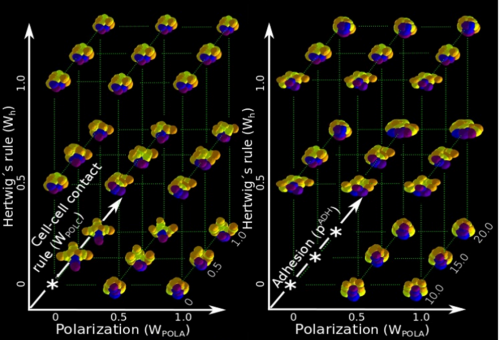
Once we had this computational model ready to use, we performed the two in-silico experiments described in the paper. First, we had to design such experiments at conceptual level (what to simulate, which phenomena to include, until which stage should one run the simulation, which data to collect, how to compare results …) because normally there is no previous experimental protocols as in wet-lab procedures. This is a rather creative stage of the project. Then, the bulk of simulations (each one taking several hours of computation time) was automatically run in order to create the theoretical morphospace. It was really exciting to look for first time the resulting simulated embryos, and to realize that they were very similar to real ones in visual appearance. This early subjective appreciation was later confirmed by quantitative measures of the morphological similarity between the real and the simulated embryos (for some species, the similarity was 100%). This was the real Eureka moment of our research !
“It was really exciting to look for first time the resulting simulated embryos, and to realize that they were very similar to real ones in visual appearance”
What next for you after this work, Miguel?
MBU I have recently presented my Doctoral Thesis (November, 2016) in the Autonomous University of Barcelona, whose main chapters were devoted to the modelling of spiral cleavage. Currently, I am looking for future (Post-Doc) projects in which I could take advantage of the abilities acquired during my PhD and that would be in consonance with my intellectual interests. In that sense, I would like to continue in Evo-Devo modeling, specially in its more theoretical side. This is because I think that this theoretical/modelling approach is the fastest, cheapest and simplest way to address many questions of capital interest in biology (e.g. how biological complexity evolves, or how development itself evolves …).
And where next for the Salazar-Ciudad lab?
ISC At the moment I have several lines of research in the lab. One is taking a similar approach that in here but in mammalian tooth development (system in which I have been working for many years). One member of my group is using EmbryoMaker (a general modelling framework developed by us, described in a previous Node post) to implement a more realistic tooth model. We have built several tooth models in the past but their bio-mechanics were not precise enough to explore in enough detail the early tooth morphogenesis. This person is also making bio-mechanical experiments on tooth germs to contrast the model predictions.
In another line of research we are using a fly wing model we published in 2015 to explore how fly morphology changes with temperature. In yet another line of research we are using an existing model of development and evolution in tooth morphology to measure how accurately the statistical approach of quantitative genetics (mostly through Lande’s equations) holds for complex multivariate morphologies that are the result of a complex developmental process.
Finally we are using the EmbryoMaker in a ensemble approach in which we build a huge number of random networks and check which embryo morphologies they produce. This apparently crazy approach has the advantage that it may allow us to identify in an unbiased way general structural principles that gene networks need to fulfil to be able to produce complex morphologies. I am also starting to look for a new place to work.
Miguel Brun-Usan, Miquel Marín-Riera, Cristina Grande, Marta Truchado-Garcia & Isaac Salazar-Ciudad. A set of simple cell processes is sufficient to model spiral cleavage. Development 144: 54-62.
Browse the People Behind the Papers archive here.


 (1 votes)
(1 votes)