Phenologs and unlikely models
Posted by Eva Amsen, on 1 September 2010
![]() “You’re probably wondering why I’m here”, were the first words of Edward Marcotte’s talk at the SDB meeting last month. After all, he was about to speak about systems biology in a session on organogenesis. What followed was not only a new way to identify genes involved in developmental processes, but also a perfect example of the kind of unexpected discoveries that can be made using publicly available data.
“You’re probably wondering why I’m here”, were the first words of Edward Marcotte’s talk at the SDB meeting last month. After all, he was about to speak about systems biology in a session on organogenesis. What followed was not only a new way to identify genes involved in developmental processes, but also a perfect example of the kind of unexpected discoveries that can be made using publicly available data.
Edward Marcotte is a bioinformatician at the University of Texas at Austin. His lab introduced the concept of phenologs to discover non-obvious disease models and candidate genes, and at the SDB meeting, as well as in a recent paper, he described exactly how “non-obvious” some of those models are: If a yeast model for angiogenesis doesn’t sound unlikely enough, the group also proposed a plant model for Waardenburg syndrome!
The concept behind phenologs is that a set of genes related to a phenotype in one organism may correspond to an orthologous set of genes in another organism. Orthologues are homologous genes between different species, but this does not necessarily mean that the same gene is linked to the same phenotype in both organisms. Marcotte looked at groups of orthologues: If a group of genes is linked to a certain phenotype in one organism, and that same group results in another phenotype in a second organism, then those two phenotypes are phenologs.
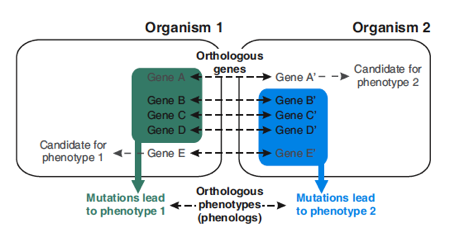
The concept of phenologs. (Figure 1B in the PNAS paper.)
In one practical example from the paper, known gene-phenotype associations from yeast were compared with known gene-phenotype associations from mice, using information from publicly available yeast and mouse genome databases. This showed that many genes that are associated with abnormal angiogenesis in mice have orthologous genes in yeast. Of course yeast doesn’t have a circulation system, so these genes can’t possibly be associated with angiogenesis in yeast, and indeed they’re not: In yeast, these same genes are involved in sensitivity to the hypercholesterolemia drug lovastatin. This suggests that lovastatin sensitivity in yeast could be a model for angiogenesis in vertebrates. To prove this, follow-up experiments showed that the transcription factor SOX13, which was identified as lovastatin-sensitive in yeast, is required for vascular development in Xenopus.
Even more surprising than finding angiogenesis genes in yeast, is that a similar comparison of phenologs suggests a plant model for Waardenburg syndrome. This disorder is caused by impaired neural crest development, and is marked by pigmentation defects and craniofacial malformations. Phenologs showed that many genes associated with Arabidopsis failing to grow in response to gravity (gravitropism) were orthologous to human genes mutated in Waardenburg syndrome, which suggests that other gravitropism genes may serve as starting points to look for other factors involved in neural crest migration.
While I was listening to this talk, I wondered whether the people who did the original yeast lovastatin screens could ever have imagined their data being used to find a new factor involved in angiogenesis. And the groups that identified gravitropism-related genes in Arabidopsis must never have thought that this could even remotely have anything to do with Waardenburg syndrome in humans! It illustrates exactly why it’s important to make data from screens and large-scale studies available to others: You often only use a small amount of the data, and buried among the rest of it is information that could be useful to people you’d never expect would benefit from it! The data in public databases speeds up research and opens up new subjects of investigation, and that is exactly why it’s there.
Kriston L. McGary, Tae Joo Park, John O. Woods, Hye Ji Cha, John B. Wallingford, & Edward M. Marcotte (2010). Systematic discovery of nonobvious human disease models through orthologous phenotypes PNAS DOI: 10.1073/pnas.0910200107


 (6 votes)
(6 votes)
Wow. Thanks, Eva.
I just found out (through the referral logs) that this post was an “Editor’s selection” on Research Blogging. I won’t add the little icon they suggest for that, because it’s a bit confusing when it just says “Editor’s Selection”, but that was really nice =)
It is interesting to study plants in order to discover human diseases.
There is questions in my mind comes while Iam reading their paper. What is their future direction for the study? In another word, what researcher spouse to do to enhance this study