From our sister journals- January 2016
Posted by the Node, on 26 January 2016
Here is some developmental biology-related content from other journals published by The Company of Biologists.

Characterisation of Slc9a6 knockout heterozygous female mice
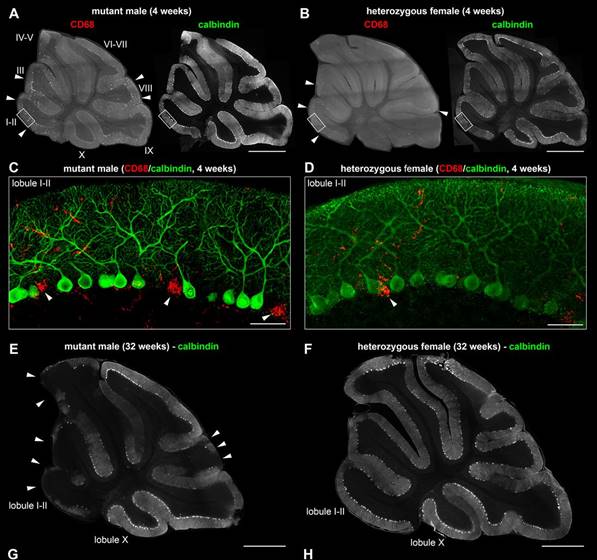 Mutations in SLC9A6 are responsible for X-linked Christianson syndrome, a neurodevelopmental disease. Sikora and colleagues demonstrate that female mice heterozygous for a Slc9a6 knockout present mosaic neuropathology and similar but milder behavioural traits to those of affected males. Read the paper here. [OPEN ACCESS]
Mutations in SLC9A6 are responsible for X-linked Christianson syndrome, a neurodevelopmental disease. Sikora and colleagues demonstrate that female mice heterozygous for a Slc9a6 knockout present mosaic neuropathology and similar but milder behavioural traits to those of affected males. Read the paper here. [OPEN ACCESS]

Sp5 emerges as an important component in mESC naïve pluripotency
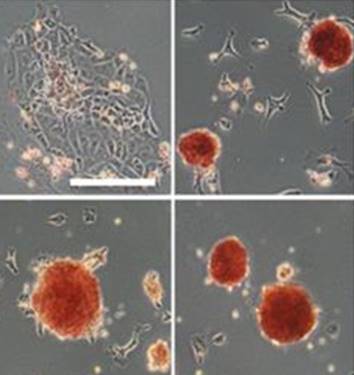 Sp5 has been identified as an effector of both Wnt/β-catenin and leukemia inhibitory factor (LIF)/Stat3 pluripotency signaling, and its forced expression produces effects of both pathways in mESC pluripotency. Furthermore, Sp5 can convert mouse epiblast stem cells into mESCs. Read the paper here. [OPEN ACCESS]
Sp5 has been identified as an effector of both Wnt/β-catenin and leukemia inhibitory factor (LIF)/Stat3 pluripotency signaling, and its forced expression produces effects of both pathways in mESC pluripotency. Furthermore, Sp5 can convert mouse epiblast stem cells into mESCs. Read the paper here. [OPEN ACCESS]
FGF signalling on E-Cadherin impacts primordial germ cell motility
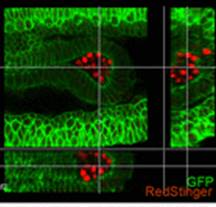 In this paper, Parès and Ricardo describe how fibroblast growth factor (FGF) signaling modulates zygotic E-cadherin distribution to maintain posterior midgut epithelial 3D architecture, impacting on primordial germ cell motility during the early embryonic development of Drosophila. Read the paper here.
In this paper, Parès and Ricardo describe how fibroblast growth factor (FGF) signaling modulates zygotic E-cadherin distribution to maintain posterior midgut epithelial 3D architecture, impacting on primordial germ cell motility during the early embryonic development of Drosophila. Read the paper here.
Targets identified for NF-κB-modulated microRNAs that inhibit myoblast proliferation
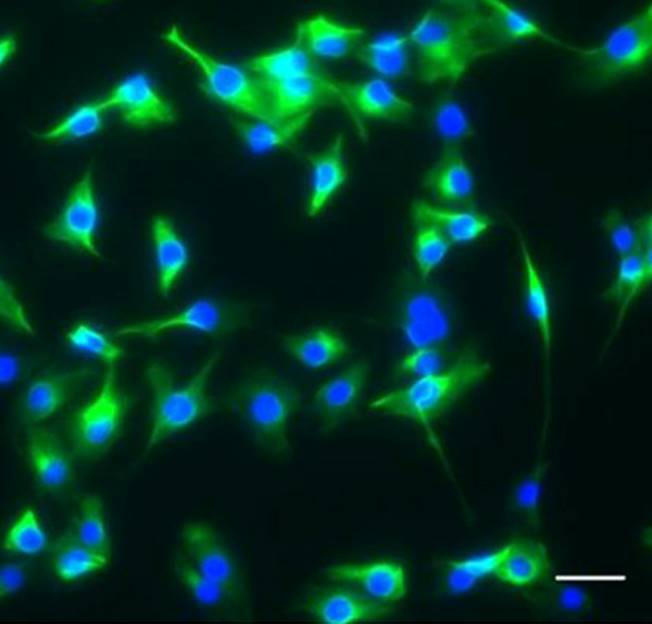 Wei and colleagues show that miR-195 and miR-497 target lgf1r, lnsr, Ccnd2 and Ccne1 and inhibit proliferation in C2C12 cells. They also show that these microRNAs are negatively regulated by nuclear factor κB, illustrating an important signalling pathway in myogenesis. Read the paper here.
Wei and colleagues show that miR-195 and miR-497 target lgf1r, lnsr, Ccnd2 and Ccne1 and inhibit proliferation in C2C12 cells. They also show that these microRNAs are negatively regulated by nuclear factor κB, illustrating an important signalling pathway in myogenesis. Read the paper here.
Reciprocal regulation of alternative lineages by Rgs18 and its transcriptional repressor Gfi1b
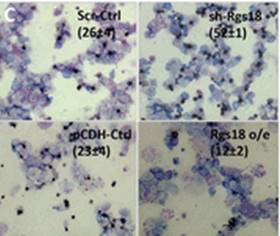 Sengupta and colleagues describe how Rgs18, a GTPase-activating protein, and its transcriptional repressor Gfi1b reciprocally regulate the lineage segregation between the megakaryocytic and the erythroid lineage through the downstream effects on the antagonistic transcription factors Fli1 and Klf1. Read the paper here.
Sengupta and colleagues describe how Rgs18, a GTPase-activating protein, and its transcriptional repressor Gfi1b reciprocally regulate the lineage segregation between the megakaryocytic and the erythroid lineage through the downstream effects on the antagonistic transcription factors Fli1 and Klf1. Read the paper here.


 (No Ratings Yet)
(No Ratings Yet)