SP7: another small step toward understanding its mechanisms of action
Posted by Julian Lui, on 7 February 2022
During embryonic development, bone formation begins with the condensation of mesenchymal stem cells (MSCs). In a few places of our body, such as in the skull and the shoulder blades, mesenchymal condensations differentiate directly into bone-forming cells called osteoblasts. These osteoblasts make bones by directly laying down bone matrix, and this process is known as intramembranous ossification. In most other condensations in the embryo, such as those that eventually became the arms and legs, bones are formed by a different process called endochondral ossification, where a cartilage mold is first formed by chondrocytes, which is then replaced by the incoming osteoblasts, laying down bone matrix using the cartilage scaffold. Therefore, osteoblast function plays an important role in both types of ossification in our body and have clinical implications in both skeletal development and diseases like osteogenesis imperfecta and osteoporosis.
Runx2 and Osterix (also known as Sp7) are two of the most well-studied transcription factors that control osteoblast differentiation. Runx2 belongs to the Runx family that is composed of three genes, Runx1/Cbfa2, Runx2/Cbfa1, and Runx3/Cbfa3. Runx2 and 3 together are essential for chondrocyte maturation (1) and Runx2 is important also for osteoblast differentiation, as demonstrated by the complete lack of ossification in Runx2 knockout mice (2, 3). The molecular mechanisms by which Runx2 regulate osteoblast differentiation has been elucidated. All Runx2 family proteins contain a DNA-binding runt domain. The Runx proteins form heterodimers with transcriptional co-activator core binding factor b (Cbfb) in vitro (4) and specifically recognize a consensus sequence, PyGPyGGTPy (5), to upregulate a variety of osteoblast lineage-specific genes, such as Sp7 (osterix), Ocn (osteocalcin), and Bsp (bone sialoprotein) (6, 7). Similar to Runx2, Sp7 knockout mice also demonstrated lack of ossification. But in contrast to Runx2 knockout, the Sp7 knockout mice do express Runx2 in osteogenic cells, thus suggesting Sp7 acts downstream of Runx2 during osteoblast differentiation (8).
Over the years, the role of Sp7 in osteoblast function has been confirmed in humans. Pathogenic variants in SP7 have been described in patients with recessive osteogenesis imperfecta (OI type XII), which is characterized by generalized osteoporosis (9), and genome-wide association studies (GWAS) have identified common genetic variants in SP7 to be associated with bone mineral density in the general population (10). However, despite the physiological and clinical importance of SP7, much is yet to be learnt about its molecular mechanisms of action. Based on its similarities with other SP family members, SP7 was initially thought to bind GC-rich sequences (11). However, other studies have shown evidence for a lack of such binding preference for GC-rich sequences (12). In 2016, a study from Andrew McMahon’s group showed that SP7 differs from other SP proteins in that it has a lower binding affinity for GC-rich sequences, and instead preferentially binds to AT-rich sequences in osteoblast target genes through interactions with DLX proteins (13). These findings provided important insights into why SP7, but not other SP family proteins with similar zinc finger domains, is uniquely important for osteoblast function. However, these findings were based on in vitro chromatin immunoprecipitation sequencing (ChIP-Seq) using osteoblasts, and thus it remains unclear whether this unique AT-motif-binding of SP7 is important in vivo, as at least until a human subject is found to have such binding preference disrupted.
And that brings us to our work recently published in Nature Communications (14). In our latest study, we presented an Austrian boy with a complex skeletal disease which included craniosynostosis, severe scoliosis, long bone fragility, with areas of increased bone, particularly with thickened intramembranous bones, and other areas of decreased bone. In that patient, we identified a pathogenic heterozygous missense variant in SP7 (S309W). The complex skeletal phenotype and the apparently dominant nature of the variant differ markedly from the prior cases of SP7-associated recessive osteogenesis imperfecta and is not readily explained by a simple loss of SP7 function and osteoblast formation.
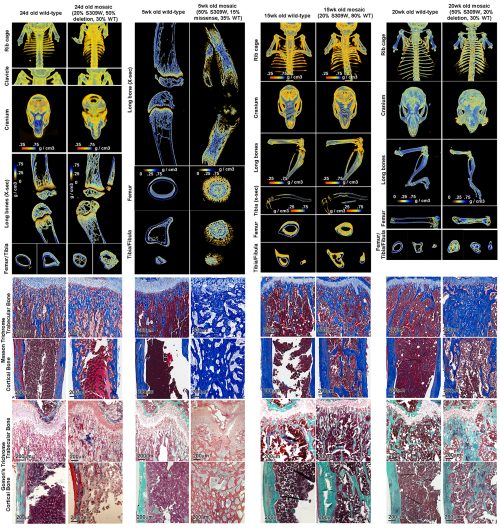
We created a mouse with the orthologous missense variant in Sp7, which partially recapitulated the human skeletal phenotype. Importantly, we showed evidence that the variant altered the binding specificity of SP7, with increased binding to GC-consensus sequences and decreased binding DLX proteins and to AT-rich motif (thus a neomorphic/gain-of-new-function mutation rather than a simple loss- or gain-of-function mutation). The variant tends to reverse the unique sequence specificity of SP7, causing it to revert to a specificity more similar to the other SP family members. Our current study therefore provides the first in vivo evidence that the unique AT-binding specificity of SP7 is indeed essential for normal bone development in vivo. Importantly, our study also suggests the possibility that other unresolved rare genetic disorders could also be caused by neomorphic mutations in transcriptional regulators.
Thank you for reading!


 (No Ratings Yet)
(No Ratings Yet)