Specifying stem cells, specifically
Posted by WIMMBlogEditor, on 5 February 2015
Bone marrow transplants save lives. It’s as simple as that.
The reason bone marrow transplants are so effective is because this squishy tissue is home to haematopoietic stem cells (HSCs), which spend their lives happily producing every single blood cell that will ever circulate around your body.
As a result, if anything goes wrong with your own blood, it is possible to remove your bone marrow (which, for whatever reason, is producing sub-optimal cells) and replace it with somebody else’s which is doing the job just fine.
But it isn’t always quite that straightforward. Finding immunologically compatible tissue isn’t trivial, and there are always more patients needing transplants than there are donors on the list. Also, even if you do manage to find a compatible donor, sometimes their bone marrow just doesn’t contain enough stem cells to be effective.
So, an alternative way of replacing the haematopoietic stem cells in patients with blood disorders is required, and has been the target of many research teams around the globe for years. The golden goal is to be able to generate haematopoietic stem cells in the lab that are genetically matched to the patient, and recent advances in generating induced pluripotent stem cells (iPSCs) from human cells has brought this prospect tantalizingly close – but to date, nobody has managed to successfully generate patient-specific HSCs in the lab.
One major stumbling block in achieving this goal has been a fundamental lack of understanding of how, when and where HSCs actually originate in the first place; how can we possibly attempt to mimic this process in the lab if we don’t know how it works in in a living, breathing organism?
This is exactly the question that scientists in Roger Patient’s lab at the WIMM in Oxford have been asking, and have published their latest findings in Nature Communications.
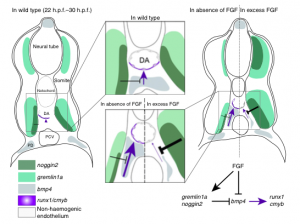
It is known that during embryonic development of all examined vertebrates, haematopoietic stem cells originate from a layer of arterial cells on the ventral side of the dorsal aorta, known as the haemogenic endothelium [1]. This process is believed to be extremely transient, and is regulated by a complex array of inputs from a variety of signaling pathways, including Wnt16 [2], VegfA [3] and Bmp4 [4].
Role of FGF signalling in the formation of HSCs. Pouget et al, Nature Comms (2014)
Recent research has shown that runx1, a key gene required for emergence of HSCs across many vertebrate species, is directly activated by the Bmp4 signalling pathway in vitro [5], and it is also known that just before definitive (or adult) haematopoiesis is activated in developing embryos, the aortic region switches from a BMP repressive to active environment [4]. However, the mechanism underlying this switch has remained elusive – until now.
Through a series of careful experiments in zebrafish, scientists in the Patient lab have found that the FGF signaling pathway is a negative regulator of HSC emergence via its control of bmp4 activity in the aortic floor, and therefore could be component of the mechanism underlying the switch from a BMP repressive to active environment that is required for HSC emergence.
Not only do these findings add a crucial piece to the puzzle in understanding how HSCs develop in vivo, but could also help to develop more efficient strategies to generate patient-specific haematopoietic stem cells in the lab.
The original research article was published in Nature Communications in November 2014: FGF signaling restricts haematopoietic stem cell specification via modulation of the BMP pathway
This post was written by Bryony Graham (@bryony_g) and was originally published on the WIMM blog.
1. Swiers, G., Rode, C., Azzoni, E., & de Bruijn, M. (2013). A short history of hemogenic endothelium Blood Cells, Molecules, and Diseases, 51 (4), 206-212 DOI: 10.1016/j.bcmd.2013.09.005
2. Clements, W., Kim, A., Ong, K., Moore, J., Lawson, N., & Traver, D. (2011). A somitic Wnt16/Notch pathway specifies haematopoietic stem cells Nature, 474 (7350), 220-224 DOI: 10.1038/nature10107
3. Leung A, Ciau-Uitz A, Pinheiro P, Monteiro R, Zuo J, Vyas P, Patient R, & Porcher C (2013). Uncoupling VEGFA functions in arteriogenesis and hematopoietic stem cell specification. Developmental cell, 24 (2), 144-58 PMID: 23318133
4. Wilkinson, R., Pouget, C., Gering, M., Russell, A., Davies, S., Kimelman, D., & Patient, R. (2009). Hedgehog and Bmp Polarize Hematopoietic Stem Cell Emergence in the Zebrafish Dorsal Aorta Developmental Cell, 16 (6), 909-916 DOI: 10.1016/j.devcel.2009.04.014
5. Pimanda, J., Donaldson, I., de Bruijn, M., Kinston, S., Knezevic, K., Huckle, L., Piltz, S., Landry, J., Green, A., Tannahill, D., & Göttgens, B. (2007). The SCL transcriptional network and BMP signaling pathway interact to regulate RUNX1 activity Proceedings of the National Academy of Sciences, 104 (3), 840-845 DOI: 10.1073/pnas.0607196104


 (1 votes)
(1 votes)