A Tale of Trunks or Zen and the art of doing a PhD
Posted by Rita Aires, on 1 September 2016
The story of this paper is also the story of my PhD. It begins as most papers and PhDs do: with a distinct and often unrelated starting project or plan. It is great to have a plan. But time and luck and data bend and twist the plan; until it finally breaks and you end up ditching said plan.
Then the real fun can begin.
I – Enter the plan
Vertebrates are an incredibly diverse group of organisms, as demonstrated by the wide range of body shapes and sizes animals belonging to this group can exhibit. This remarkable flexibility was likely a decisive factor allowing for the amazing adaptation of vertebrates to almost all habitats throughout the globe. The general body plan of a vertebrate is usually determined in embryonic development during the process of axial extension, in which the embryo elongates in a rostral to caudal progression by the activity of a very particular population of cells located in the posterior embryonic end. These are the axial progenitors, and their job is to proliferate and give rise to all post-cranial structures of the embryo (Wilson et al., 2009). Yet, an in-depth study of this important cell population was – and still is – hindered by the lack of specific molecular markers.
My initial project was then, to do a molecular characterization of axial progenitors in the mouse embryo; and my supervisor, Moisés Mallo, and I had a nice, neat strategy designed to fish these markers out. At the same time – like a responsible and efficient (and somewhat neurotic) PhD student – I had compiled quite a list of candidate genes that might be expressed and/or could potentially be important to axial progenitors just by searching for truncation phenotypes in existing databases. This way I could screen those genes while waiting for the other experiment’s results. One of these candidate genes was Oct4.
II – How I met Oct4
Oct4 is the gene equivalent of a rock star. Besides being an integral part of the core pluripotency network, Oct4 is also famously known for being an indispensable part of the reprogramming cocktail able to turn somatic cells back into pluripotent stem cells (Takahashi and Yamanaka, 2006). All vertebrate species studied so far seem to have a functional Oct4 counterpart (Frankenberg et al., 2014). In the mouse embryo, Oct4 is expressed as early as the two-cell stage, becoming gradually down regulated as development progresses until its complete silencing in somatic tissues by mid-gestation stages. However, in 2013, a paper came out by DeVeale and colleagues (DeVeale et al., 2013), which showed that, if Oct4 was inactivated during a very particular window of developmental time, some of the resulting mouse embryos could exhibit significant axial truncations, while others showed dramatically shortened trunks, yet perfectly specified tails. The latter were remarkably similar to a variety of transgenic embryos we had described just a few months before, in which the type I TGF-β receptor Alk5 was constitutively active in axial progenitor-containing areas (Jurberg et al., 2013). This showed that not only Oct4 seemed to be important for trunk development, but also suggested the existence of some sort of inverse functional link between Oct4 expression and TGF-β signalling through Alk5 during mouse axis elongation.
But was that really true? At that point, I and my good friend and former labmate Arnon D. Jurberg had been working with this pathway for quite some time, specifically with the Gdf11 mutant. Gdf11 is the main ligand for the Alk5 receptor in vivo, and mutants for both these proteins have characteristically longer trunks due to a delayed trunk to tail transition (Andersson et al., 2006; Jurberg et al., 2013; McPherron et al., 1999). So we definitely knew that these molecules were important for axial progenitors during the extension process. Thus, we decided to put this putative functional connection to the test using our faithful Gdf11 mutant. We reasoned that if premature activation of Alk5 activity was equivalent to the absence of Oct4 during axis formation, and this excess signalling was able to generate shorter trunks, then perhaps the lack of signalling through Alk5 – by absence of Gdf11, for example – could result in an increased expression of Oct4 and that could maybe account for the longer trunks in these mutants. Admittedly, it was a bit of a stretch. But to our vast surprise and amazement, our preliminary results showed us that we might actually be right.
III – Oct4 and the “snakefied” mice
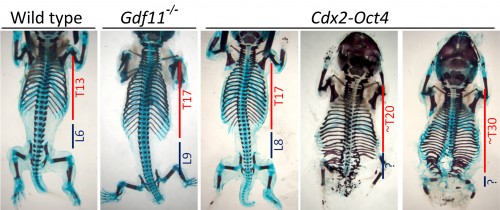
Proving beyond any doubt that Oct4 was really and truly ectopically expressed in Gdf11 mutants took sensibly two years, three different methodologies and a healthy dose of stubbornness. However, in the meantime, we still wanted to know if Oct4 missexpression was responsible for the longer trunks these mutants showcased, which required a way to overcome its progressive downregulation throughout development. Therefore, we chose to take advantage of the Cdx2 promoter, which we had previously demonstrated to be active in axial progenitors, and use it to drive Oct4 expression in transgenic mouse embryos. So it was a dark December day – and a national holiday to boot – when Moisés and I were lucky to dissect together a particularly good batch of E18.5 transgenic embryos courtesy of Ana Nóvoa, transgenic wizard extraordinaire. And the results surpassed even our wildest expectations. Not only were we able to obtain embryos that mimicked the Gdf11 mutant axial phenotype, we also got fetuses that yielded up to 20 or 30 trunk segments (Fig.1). Which meant two things: first, that Oct4 missexpression was most likely the main factor responsible by the longer trunks in Gdf11 mutant embryos; and second, that the trunk to tail transition could be delayed almost indefinitely if you just maintained the right level of Oct4 activity during axial extension.
IV – How the snake got its curves
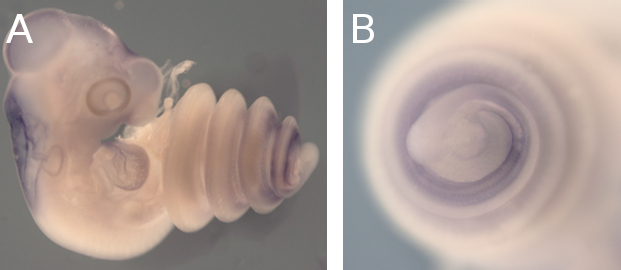
Needless to say, if something has long, rib-packed and organ-filled trunks… well, it’s snakes. Could Oct4 have a role in the making of their long trunks too? For that we needed to check its expression during snake embryonic development, which was most definitely not our area of expertise. We decided to ask for help from Francisca Leal and Martin J. Cohn from the University of Florida, proud owners of a lovely snake facility and source of infinite wisdom in all snake-related matters. We hit the jackpot: expression studies revealed that Oct4 was still present in snake embryos long after it was gone in equivalently staged mice embryos (Fig.2). That made for a classic case of gene expression heterochrony, in which the timing of a gene’s activation or silencing changes from one species to another.
What could have possibly happened during evolution for Oct4 to be so differently regulated in these two groups of organisms? Thanks to Francisca and her sequence analysis skills, we found out that the genomic landscape upstream of the snake Oct4 locus had diverged dramatically from mice and even lizards. Yet, this entire region was extremely well conserved among snakes. As sequence conservation generally means conservation in function, we wondered if these conserved non-coding sequences 5’ of snake Oct4 had any regulatory potential by testing them in mice. We found that a tiny, little 250bp region – the only region conserved in both lizard and snake species – was extremely active on its own, whereas when embedded within larger snake-specific sequences it was a lot less able to drive reporter gene expression. But these were only scattered, isolated pieces of a bigger puzzle. When we used a snake BAC containing the whole genomic context around Oct4 – a kind gift from Isabel Guerreiro and Denis Duboule – and got no expression in transgenic mice, we realized that this entire region most likely operated as a complex regulatory module, composed by a 250bp core regulatory enhancer under the additional control of neighbouring sequences. In the end, we think that the remarkable conservation in snake non-coding sequences probably resulted from strong selective pressures over Oct4 expression due to the adaptation to a fossorial lifestyle, which favoured long trunks for burrowing and prey constriction.
V – Epilogue
So there you have it: the story of how ditching the initial plan can be a good thing, if you just pay attention and let it happen. Especially when results start acquiring a life, to tell a story of their own. Go with the flow. Get really curious and excited. Talk to people, ask for help, and help out even more. And maybe, just maybe, you might stumble across something that, ultimately, turns out to be your dream Evo-Devo project.
References
Aires R, Jurberg AD, Leal F, Nóvoa A, Cohn MJ, Mallo M. (2016) Oct4 Is a Key Regulator of Vertebrate Trunk Length Diversity. Dev Cell. 2016 Aug 8;38(3):262-74
Andersson, O., Reissmann, E. and Ibáñez, C. F. (2006). Growth differentiation factor 11 signals through the transforming growth factor-beta receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep. 7, 831–7.
DeVeale, B., Brokhman, I., Mohseni, P., Babak, T., Yoon, C., Lin, A., Onishi, K., Tomilin, A., Pevny, L., Zandstra, P. W., et al. (2013). Oct4 Is Required ˜E7.5 for Proliferation in the Primitive Streak. PLoS Genet. 9, e1003957.
Frankenberg, S. R., Frank, D., Harland, R., Johnson, A. D., Nichols, J., Niwa, H., Schöler, H. R., Tanaka, E., Wylie, C. and Brickman, J. M. (2014). The POU-er of gene nomenclature. Development 141, 2921–3.
Jurberg, A. D., Aires, R., Varela-Lasheras, I., Nóvoa, A. and Mallo, M. (2013). Switching Axial Progenitors from Producing Trunk to Tail Tissues in Vertebrate Embryos. Dev. Cell 25, 451–462.
McPherron, A. C., Lawler, A. M. and Lee, S.-J. J. (1999). Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22, 260–264.
Takahashi, K. and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676.
Wilson, V., Olivera-Martínez, I. and Storey, K. G. (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 2133–2133.


 (8 votes)
(8 votes)