Testing constructs for CRISPR/Cas9 genetic modulation
Posted by Merissa Hickman, on 11 October 2021

This summer, I was given the opportunity to conduct research at the Francis Crick Institute in the Znamenskiy lab. The aim of the Znamenskiy lab is to understand the relationship between connectivity, gene expression and function of cortical neurons.
The neocortex is a region of the brain integral in performing higher cognitive functions. Neocortical projections can be divided into three broad classes. Corticothalamic (CT) neurons are located mostly within layer 6 and send axons to the thalamus. Pyramidal Tract (PT) neurons are nearly exclusively positioned within layer 5 and project to brainstem and spinal cord. Intratelencephalic (IT) neurons are distributed throughout all six layers and project to distant cortical areas (Kast & Levitt, 2019). The expression of transcription factors during development can affect projection patterns. For example, when Fefz2 is deleted, the cortex no longer sends projections to the brain stem and instead sends projections to the thalamus or contralateral hemisphere (Kast & Levitt, 2019). This shows that genes expressed by a neuron during development play an important role in determining its wiring patterns.
Beyond these broad projection classes, the genetic basis underlying patterns of neocortical connectivity is little understood. The primary visual cortex (V1) is a region of the brain that is important for receiving, segmenting, integrating, and processing visual information relayed from the retinas. Subsequently, the processed information is then sent to other regions of the brain. This is a highly specialised process that allows the brain to recognise patterns quickly and with the absence of a conscious effort. The V1 provides a platform for understanding the neocortex due to its serially homologous structure, and therefore can be used as a model for neocortical projections. The V1 sends projections to several higher visual areas as well as many other areas of the brain such as the lateral geniculate and lateral posterior thalamic nuclei, superior colliculus, striatum, and other subcortical structures but little is known about how these connectivity patterns are established (Kast & Levitt, 2019).
To understand which genes are important for specifying long range connectivity patterns from V1, in vivo genetic manipulations using CRISPR/Cas9 can be used to determine what happens to connectivity patterns when the expression of target genes is altered. CRISPR/Cas9 is a simple, rapid method to modify gene expression which can be pooled together to look at many genes in parallel. As well as knocking-out the gene of interest using the prototypical CRISPR/Cas9 gene editing approach (Figure 1A), methods for modulating gene expression using catalytically inactive Cas9 fused to transcriptional modulators have recently been developed (Figure 1B-C). CRISPR activation (CRISPRa) allows functional analysis of redundant genes through overexpression, whereas CRISPR interference (CRISPRi) allows analysis of gene function by knocking-down gene expression at the transcriptional level and is thought to have fewer off-target effects (Gebre et al., 2018). The aim of my project was to perform preliminary experiments validating whether gRNA constructs designed to be used to examine changes in in vivo V1 connectivity patterns, using CRISPR knockout, CRISPRi or CRISPRa, altered gene expression in vitro. The first part of my project was to clone some of the gRNA CRISPR constructs, and the second part was to test constructs in vitro.
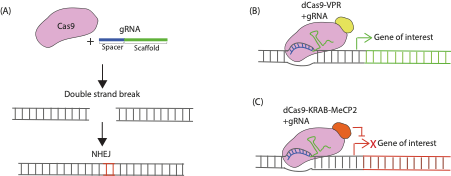
FIGURE 1. Mechanism of CRISPR/Cas9 Genetic Modulation. (A) CRISPR knockout involves co-expressing Cas9 and a gRNA in a cell. The Cas9 protein recognises a specific sequence called the scaffold sequence in the gRNA while another sequence within the gRNA called the spacer region determines the target site within the genome to be modified. The Cas9 protein generates double strand breaks in the gene of interest that are repaired through the non-homologous end joining (NHEJ) pathway that is prone to producing indel mutations (red bases here indicate an insertion) that can render genes non-functional when translated. (B) CRISPR activation (CRISPRa) constructs work via transcriptional activators fused to catalytically dead Cas9 (dCas9) which are targeted near transcriptional start sites of the endogenous gene of interest by the gRNA to induce their overexpression. (C) CRISPR interference (CRISPRi) constructs consist of dCas9 fused to transcriptional repressor domains that are recruited in proximity of the transcription start site of an endogenous gene to repress transcription.
gRNA constructs were tested along with corresponding Cas9s (SP-Cas9, dCas9-KRAB-MeCP2, and dCas9-VPR for CRISPR knockout, CRISPRi, and CRISPRa, respectively) to determine whether a change of expression in our genes of interest occurred within Neuro-2A (N2a) cells. Target genes for validation (Frizzled 1 (FZD1), Androgen Receptor (AR), Polycystic Kidney and Hepatic Disease 1 (PKHD1), and Anaplastic Lymphoma Kinase (ALK)) were identified due their established endogenous gene expression in N2a cells. To determine whether the gRNA constructs worked I co-transfected Cas9’s with the gRNA construct into N2a cells and observed whether this altered expression of target genes by looking at endogenous protein levels through immunostaining. Endogenous protein levels in each condition were compared to a control plasmid without a gRNA insert. The results obtained from the quantification of the transfection and subsequent immunostaining are shown in Figure 2A-C. These results did not reveal expected differences in gene expression between gRNA constructs and further experiments need to be performed using alternative antibodies or staining conditions. However, the project has given me an insight into the molecular basis of developmental biology, and I thoroughly enjoyed learning the techniques and protocols required to complete the cloning process. During my research internship I was able to obtain applied, practical experience within the laboratory which due to the COVID-19 pandemic, has been limited during my undergraduate degree. I also was given a level of independence which I did not expect within the laboratory, completing the transfection of gRNA constructs was an engaging, albeit challenging process as my cells became contaminated during the passaging process. However, I was able to overcome this setback and build resilience. Overall, I really enjoyed my project, and it has encouraged me to pursue a career in scientific research.
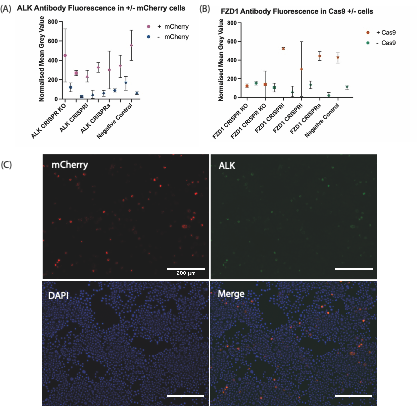
FIGURE 2. Quantitative Immunofluorescence after transfection of Cas9 plasmids and mCherry expressing gRNA plasmids targeting either ALK or FZD1. (A) ALK antibody fluorescence in +/- mCherry Cells. (B) FZD1 antibody fluorescence in + mCherry; +/- Cas9 Cells. KO/i/a – KO = Knockout; i= interference, a = activation. Each symbol shows the mean normalized grey value of N2A cells which reflects the level of fluorescence from antibodies targeting the endogenous protein-of-interest after immunostaining. The negative control used was a plasmid without a gRNA insert. SP-Cas9 was used for ALK/FZD1 KO & negative control, whereas dCas9-KRAB-MeCP2 was used for ALK/FZD1 CRISPRi, dCas9-VPR used for ALK/FZD1 CRISPRa. For the FZD1 transfection there was an unexpectedly low number of +Cas9 cells. (C) Immunohistochemistry staining against mCherry and ALK, as well as DAPI staining in N2a cells transfected with SP-Cas9a and a gRNA targeting ALK. mCherry is expressed by the gRNA constructs, staining this protein shows which cells were transfected with our construct of interest, whereas DAPI staining marks the nuclei of all cells. The overlap in ALK and mCherry signals suggests further optimisation of immunostaining and imaging conditions is required to avoid bleed-through.
I would like to take this opportunity to thank the Francis Crick Institute, particularly the Znamenskiy lab for allowing me to undertake research at their facility, alongside my supervisor Benita Turner-Bridger for supporting me in my project. Furthermore, I would like to show my appreciation to the Medical Research Foundation and the Rosa Beddington fund which has provided the financial support for my project. It is an honour to have the opportunity to contribute to The Node and the British Society of Developmental Biology, and I would strongly encourage other undergraduate students to pursue a similar research project during their studies. This experience has been unlike any other.
References:
Gebre, M., Nomburg, J.L and Gewurz, B.E (2018). CRISPR-Cas9 Genetic Analysis of Virus-Host Interactions. Viruses, 10(2), 55.
Kast, R.J and Levitt, P (2019). Precision in the development of neocortical architecture: From progenitors to cortical networks. Progress in Neurobiology, 175, 77-95.


 (2 votes)
(2 votes)