The people behind the papers – Kenji Nagata and Mitsutomo Abe
Posted by the Node Interviews, on 28 June 2021
This interview, the 93rd in our series, was published in Development earlier this year.
The plant epidermis is a single layer of cells that forms a crucial barrier to the outside world, but the mechanisms that control epidermal differentiation – in particular the relative importance of position and lineage – remain incompletely understood. A new paper in Development tackles this question in Arabidopsis. To find out more about the story, we caught up with first author Kenji Nagata and his supervisor Mitsutomo Abe, Associate Professor at the University of Tokyo.

Mitsutomo, can you give us your scientific biography and the questions your lab is trying to answer?
MA: As a PhD student I researched molecular genetics in Arabidopsis in the lab of Yoshibumi Komeda at Hokkaido University in Sapporo, Japan. I was excited to go to the lab every day, and so fascinated by the beautiful expression patterns of ATML1 and PDF2, twin genes that enabled me to get my PhD in 2001. After my PhD, I joined the lab of Takashi Araki at Kyoto University as an Assistant Professor and started the ‘florigen quest’ with great colleagues. I was fortunate to make a fundamental discovery regarding florigen in 2005, and I am very pleased that FT is now well known as an important component of florigen signalling. After this, I had a great experience working in the lab of Richard Amasino at the University of Wisconsin for two years, and very much enjoyed American life. I then moved to the University of Tokyo and started my own research group in 2009, and keep working on plant molecular genetics.
Our research group has a broad interest in plant development. Some members are involved in elucidating florigen function in Arabidopsis, others (like Kenji) are working on epidermal cell differentiation, and others are interested in the interaction between meristem identity and plant architecture. All have one thing in common: we are focusing on phenomena and molecules that are unique to plants.
Kenji – how did you come to work in Mitsutomo’s lab and what drives your research today?
KN: When I was an undergraduate student, I was very interested in two aspects of sessile plants: the phenotypic plasticity they show in morphology and physiology, and their developmental robustness in terms of patterning. So I looked for a lab that would allow me to explore these issues. I was fortunate to attend one of Mitsutomo’s lectures, where he described his lab’s work on flowering, which is mediated by signals from the external environment, and robust protodermal cell differentiation, which is not influenced by the external environment. I felt that working with him would provide me exciting research opportunities, and decided to join his lab. Curiosity about how the traits unique to sessile plants work and why they developed in an evolutionary context drives my research today.
How has your research been affected by the COVID-19 pandemic?
KN: Due to entry restrictions to the university, we were forced to suspend our experiments and stay at home. Fortunately, none of the members of our lab have caught the virus and all have stayed healthy so far. Although now there are still some restrictions, it is slowly getting back to (a new) normal.
MA: Like most universities and research institutes, only the minimum number of staff necessary to maintain plants and equipment was permitted to enter the lab from spring to summer. Since mid-July, research activities are allowed with the utmost care to prevent the spread of infection. But the number of infected patients in Tokyo is increasing recently, so I’m worried that the lab will be closed again.
Before your work, what was known about the relative roles of lineage and position in plant epidermis differentiation?
KN: It is widely accepted that cell position rather than cell lineage is important for plant cell fate decisions. For example, the inner cells, which derive from occasional periclinal divisions of epidermal cells, develop according to their position rather than their epidermal cell lineage. On the other hand, it is also known that, in shoots, the inner cells never adapt their epidermal identity even if they occupy the outermost position, suggesting that cell lineage is involved in the epidermal cell differentiation. However, it was recently shown that when cells in the inner cell lineage are displaced to the outermost position through laser ablation, they appear to acquire root epidermal cell fate. Thus, it is controversial whether cell lineage indeed affects epidermal cell fate decisions.
Can you give us the key results of the paper in a paragraph?
KN: In this paper, we found that ATML1, a master regulator of protoderm/epidermis differentiation, is only stabilized in the outermost cells derived from the outermost cell lineage. Furthermore, the stability of ATML1 in these cells is conferred by the interaction with its lipid ligand VLCFA-Cers. VLCFA-Cers appears to be polarly localized to peripheral domains in epidermal cells, and passed on to the outermost epidermal cells in a cell position- and lineage-dependent manner. Based on these results, we have proposed a novel model in which ATML1-VLCFA-Cers interaction is restricted to the outermost epidermal cells and consequently restricts protoderm/epidermis differentiation to the appropriate position.
MA: I think for me the key experiment in our paper is the transient induction of ATML1 by a heat shock treatment – I was very excited when Kenji first came to show me the GFP images.
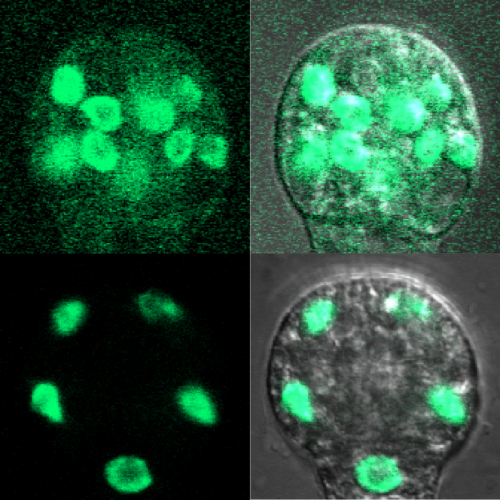
When outermost cells divide asymmetrically, the inner cells inherit ATML1 protein: what then stops them from differentiating as protoderm?
KN: This is because the ATML1 protein in inner cells is rapidly broken down, as shown in the careful observation in the 16-cell-stage embryo of gATML1-EGFP plants, or in our transient ectopic expression assay using HSP::NLS-mCherry; HSP::ATML1-EGFP plants in which the inner cells are not able to differentiate into protoderm. The rapid breakdown of ATML1 protein is due to the absence of VLCFA-Cers, in inner cells. Thus, we propose that VLCFA-Cers act as a landmark of the outermost cell position and lineage, and act as a post-translational signal that mediates positional information.
MA: Epidermis-specific expression of ATML1 and PDF2 is strictly regulated. Therefore, in addition to the mechanism we reported here, I believe that several other regulatory mechanisms are involved in epidermal cell differentiation. I’m really looking forward to the seeing this research progress in the future.
I was very excited when Kenji first came to show me the GFP images
Do lipid-transcription factor complexes mediate positional signals elsewhere in plant development?
KN: We would assume so. Kathrin Schrick and colleagues have shown that START domains from plant HD-Zip transcription factors bind lipid ligands to regulate transcription factor activity in a yeast system. Together with our findings, this suggests that lipids may mediate positional signals and modulate HD-Zip transcription factor activity elsewhere in plant development.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
KN: I think the most memorable moment was when I observed the outermost cell-specific ATML1-EGFP signal after heat-pulse treatment. This was an important moment when I was certain that our hypothesis was right.
And what about the flipside: any moments of frustration or despair?
KN: The purification of the START domain as a soluble form was challenging. I first tried to purify the full-length ATML1 protein as a soluble form, but I couldn’t obtain it, and ultimately it took over a year to obtain the soluble START domain.
What next for you after this paper?
KN: I am interested in how the ATML1 protein is broken down when VLCFA-Cers is absent – this will deepen our insights about lipid-mediated modulation of transcription factor activity in plants. On the other hand, from an evolutionary perspective, it is important to know whether lipid-transcription factor-based developmental mechanisms also work in the basal land plants or algae.
Where will this story take the Abe lab?
MA: As I mentioned in my biography, since my PhD I’ve been very interested in the molecular mechanism of epidermal cell differentiation. For the past 10 years or so, my lab has been focusing on florigen function and regulation, which involves FT, FD and FE. But I am very pleased that Kenji was interested in and restarted this project 5 years ago. In the future, Kenji and I hope to make exciting discoveries on epidermal cell differentiation.
Finally, let’s move outside the lab – what do you like to do in your spare time in Tokyo?
KN: I love to spend my spare time at the Onsen (a Japanese hot spring). After Onsen, I always drink a beer!
MA: In Spring 2019, our lab moved from Hongo to Komaba, which is a 20-min walk from Shibuya city. Shibuya city is a centre for modern culture and entertainment in Japan. I hope I will get to properly enjoy Shibuya life after the COVID-19 pandemic has settled down.


 (No Ratings Yet)
(No Ratings Yet)