Transposons in Embryo Space – TRACER maps in EMAGE
Posted by chris.armit@igmm.ed.ac.uk, on 9 February 2017
A recent publication in Developmental Biology by (Armit et al., 2017) describes how the TRACER dataset can be spatially compared with in situ hybridisation gene expression profiles.
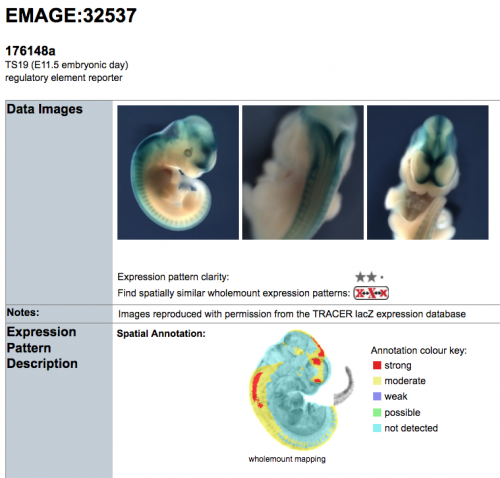
- The TRACER dataset of transposon-associated regulatory sensors (Chen et al., 2013) utilises Sleeping Beauty lacZ transposons that have been randomly integrated into the mouse genome
- Hundreds of insertions have been mapped to specific genomic positions, and the corresponding regulatory potential is documented through lacZ imaging of E11.5 wholemount mouse embryos
- Through spatial mapping of the lacZ expression patterns, the EMAGE gene expression database enables co-localisation and co-expression of regulatory elements to be explored computationally
- Spatial mapping additionally enables rapid identification of cis-regulatory elements that are expressed in a region of interest in the mouse embryo
Click here to access the spatially mapped TRACER dataset in EMAGE.
References
Armit C, Richardson L, Venkataraman S, Graham L, Burton N, Hill B, Yang Y, Baldock RA. eMouseAtlas: An atlas-based resource for understanding mammalian embryogenesis, Developmental Biology, Available online 2 February 2017, ISSN 0012-1606, http://dx.doi.org/10.1016/j.ydbio.2017.01.023
Chen, C-K, Symmons O, Uslu VV, Tsujimura T, Ruf S, Smedley D, Spitz F. TRACER: a resource to study the regulatory architecture of the mouse genome. BMC Genomics 14 (2013), p. 215, https://dx.doi.org/10.1186/1471-2164-14-215


 (No Ratings Yet)
(No Ratings Yet)