Colouring the Mouse Embryo
Posted by wongmi22, on 13 August 2012
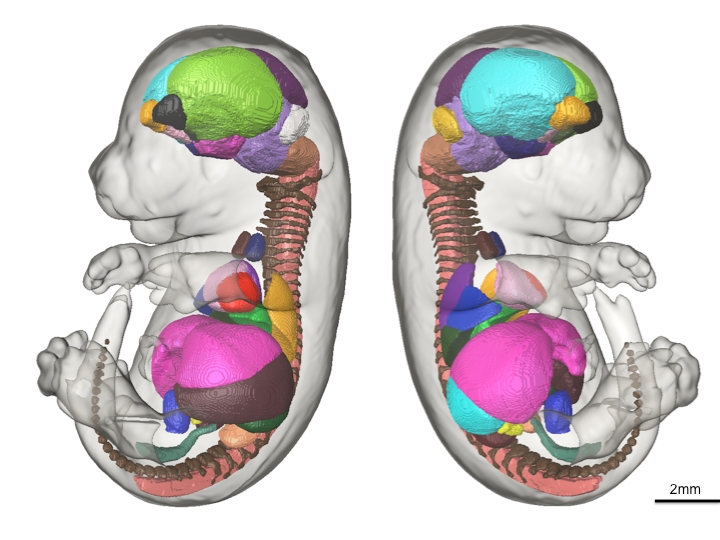
48 anatomical structures of the presented mouse embryo atlas are shown in 3D.
Here is the backdrop for our recent paper in Development, “A novel 3D mouse embryo atlas based on micro-CT”. With the human genome project complete, the sequence and the location of each gene in the genome is understood. However, the understanding of gene function and the corresponding expressed phenotype for all the genes in the human genome is still in its infancy. Most of the research aimed to tackle this question will be carried out in the mouse due to the 99% genetic homology between mice and humans and the available techniques to manipulate mice genetically. Over the last decade, the efforts of a world wide consortium, the International Knockout Mouse Consortium (IKMC, www.knockoutmouse.org), has embarked on a mission to knock out each of the ~23,000 genes in the mouse genome, one at a time, and generate the resultant mice. With this effort now close to completion another world-wide effort, the International Mouse Phenotyping Consortium (IMPC, www.mousephenotype.org), has been established and the plan on how to phenotype the resultant mice from the IKMC project is being formulated. What is well understood is that ~30% of the gene knockout mice strains will be embryonic lethal, further accentuating the need for an assay to phenotype mouse embryos throughout development.
If you have two groups of mouse embryos, one wild-type and one mutant, with a single gene knockout, how do you find out what’s different about them? How do you get clues to the function of the knocked out gene and its role in mouse embryo development? The most intuitive answer would be to look at the two groups of mouse embryos with a microscope and see if you can find any gross differences in morphology in the mutant group. You could hypothesize that the organ or structure that shows an aberration in comparison with the wild-type group is an area where that particular gene function is important and carry on with more focused phenotyping assays from there.
This is the exact premise of our recent paper in Development. Our aim was to eliminate the human bias and time needed to parse through thousands of high-resolution images by developing automated computer methods that could export volume measures of each of the major organ structures within the mature mouse embryo. We used advanced high-resolution 3D imaging called Micro-CT to image 35 E15.5 C57/Bl6 mouse embryos and developed sophisticated computer software to automatically calculate the mean volumes and standard deviation of 48 structures inside the mouse embryo. To achieve this, each of the 48 structures within a representative average image of all 35 mouse embryo images were manually painted by one individual, totalling ~400 hours of work. Through this we acquired baseline volumetric measurements of wild-type mice to determine how tight the variation is among controls. The resulting labeled data set (the above figure) exists as an E15.5 mouse embryo atlas for which all future mutant strains can be compared with and automatic volume measurements can be executed. The results presented in this paper, in our opinion, is an important step in demonstrating the feasibility of using 3D imaging as a primary screen in the IMPC pipeline and provides a robust tool that can handle and analyze the large volume of images that will be acquired.
![]() Wong MD, Dorr AE, Walls JR, Lerch JP, & Henkelman RM (2012). A novel 3D mouse embryo atlas based on micro-CT. Development (Cambridge, England), 139 (17), 3248-56 PMID: 22872090
Wong MD, Dorr AE, Walls JR, Lerch JP, & Henkelman RM (2012). A novel 3D mouse embryo atlas based on micro-CT. Development (Cambridge, England), 139 (17), 3248-56 PMID: 22872090


 (11 votes)
(11 votes)