Lighting Up the Central Dogma in Development
Posted by Garcia Lab, on 19 June 2018
We recently published a manuscript in Cell that describes a method to image transcription factor concentration dynamics in real time, in living embryos, using a nanobody-based protein tag that we call the “LlamaTag.” We were particularly excited about these investigations because this new technology overcomes a major technical obstacle to understanding how gene-expression dynamics are regulated in development, which has held back the field for decades.
From Snapshots of Dead Embryos to Movies
Remember when you were first starting to study developmental biology? In one of your lab sections, you probably participated in that classic teaching activity where you watched the cephalic furrow form in Drosophila embryos. Maybe you were one of those students struggling to move embryos with an eyelash glued to a toothpick, and the whole time you were thinking, “I need to hurry up—this thing is gastrulating while I’m messing around here!” There’s a strong contrast between watching development unfold before your eyes and scrutinizing snapshots of FISH data from dead, fixed embryos. This contrast hinders more than budding scientific excitement: how can we truly understand dynamic processes by analyzing static data?
I was particularly struck by this conundrum as a postdoctoral researcher as my mind kept replaying a video of a gastrulating embryo. How could I bridge this gap so that developmental biologists could accurately model—and ultimately construct—dynamic systems as complex as embryos? At the simplest level, we would need to know the input and the output of each genetic circuit as it changes throughout development in order to deduce the logic gates at play. I tackled the output end first: as a postdoc, I developed a technique to quantify transcription in flies at the single-cell level by tagging a gene’s nascent mRNA molecules with fluorescent proteins. How could we accomplish the same feat with the DNA-protein interactions that constitute the input to these genetic circuits? I decided that solving this challenge would be a major driver of the research in my own lab. Jacques Bothma was equally excited about this challenge and decided to join the project as a postdoctoral fellow.
Typically, one would quantify the concentration of a transcription factor by fusing it to a fluorescent protein. However, in flies, worms, fish, and frogs, these fluorescent take more than 40 minutes before they mature and become fluorescent. This delay is actually a major problem: the activators and repressors that drive development often exist for less than 10 minutes before they are degraded. By the time the fluorescent proteins actually became fluorescent, the action they were supposed to report on is already over!
Jacques had a great idea: instead of relying on the synthesis of fluorescent proteins, why not use the localization of already matured fluorescent proteins to report gene expression? Our Cell paper describes the engineering and implementation of this idea with LlamaTags, a technology that enables quantitation of the dynamics of transcription-factor activity without limits from the slow maturation of traditional fluorescent fusion proteins. Instead of these fusions, we employed nanobodies, which are small, highly specific, single-domain antibodies that are raised in llamas (hence LlamaTags). We fused a transcription factor of interest (we started with Hunchback) to a nanobody raised against eGFP, and expressed the construct from the endogenous locus for that transcription factor. Importantly, the embryo was engineered to contain maternally deposited eGFP, which means that eGFP is already mature before the transcription factor of interest is expressed (no waiting around for the fusion to mature!). When the transcription factor-nanobody is translated, it binds cytoplasmic eGFP within seconds, yielding a quantitative increase in nuclear fluorescence when the transcription factor moves to the nucleus to perform its regulatory function. LlamaTags therefore deliver a direct readout of the instantaneous transcription-factor concentration in a given nucleus.
Through a variety of experiments, we showed that LlamaTags serve as specific and faithful reporters of the endogenous concentration dynamics of transcription factors during development, thus capturing the input pertinent to these circuits. So, we finally had the two pieces needed to solve the puzzle of measuring input-output functions. LlamaTags made it possible to measure the input concentration of transcription factors in individual nuclei, while MS2 revealed the transcriptional activity of specific genes to these input levels.
By fusing a LlamaTag to Fushi-Tarazu (Ftz), we obtained measurements of its nuclear concentration during development, from which we extracted the in vivo degradation rate of the construct. We were pleased when our data revealed rapid fluctuations in Ftz concentration—consistent with previous reports of protein bursts that likely arise from stochastic fluctuations in mRNA concentrations. We nailed down this relationship, and uncovered exciting evidence of inter-nuclear communication within the embryo. This coupling could be a major driver of the sharp boundaries that dictate development of the fly embryo. However, our greatest satisfaction came from simultaneously visualizing input transcription-factor activity and output transcription, at the single-cell level, in real time, as stripe 2 of eve was laid down in live embryos:
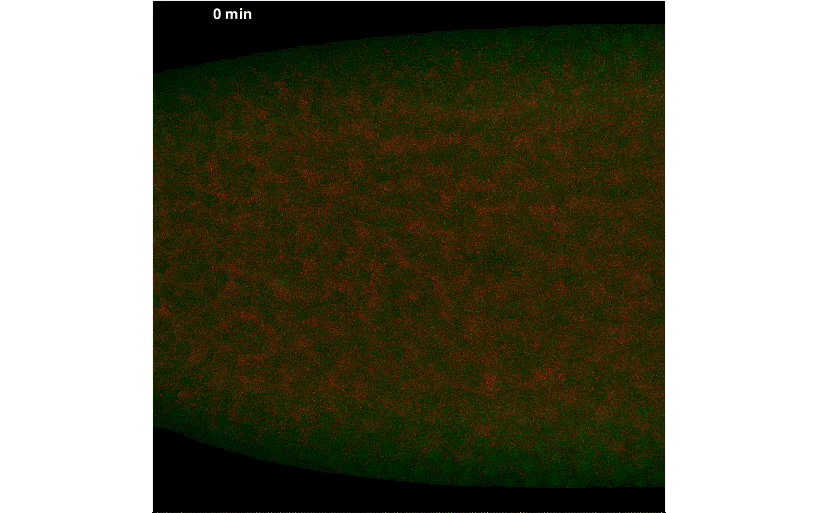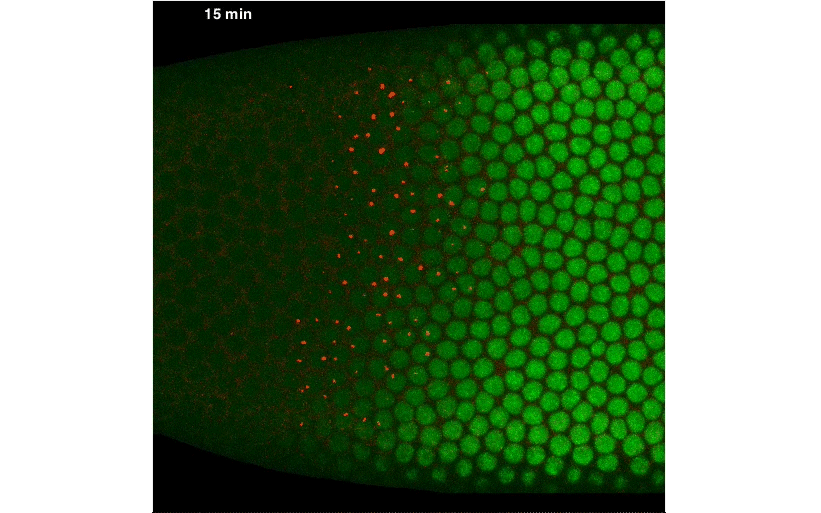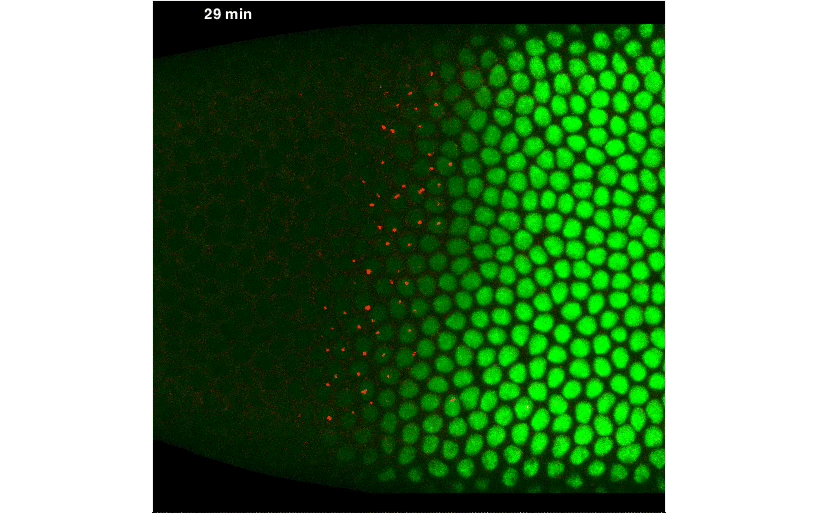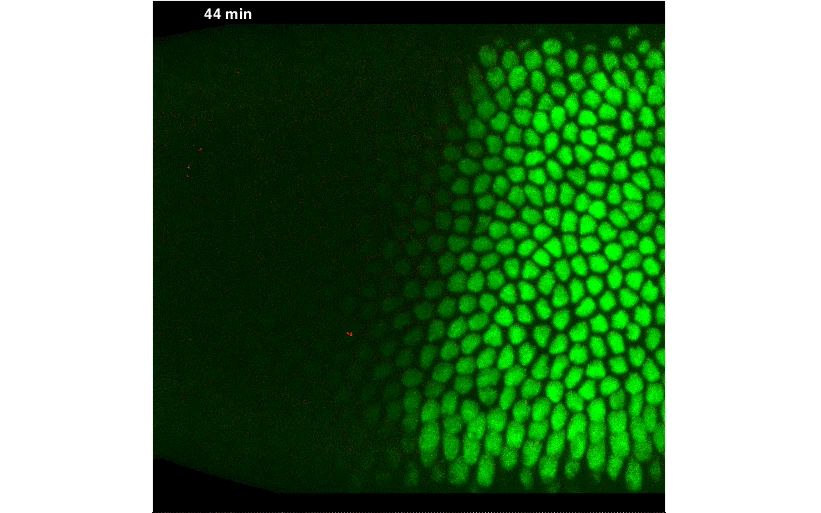
A New “Microscope” for the Central Dogma in Development
Our recent work establishes a powerful pair of technologies—labeling transcription with MS2 and labeling DNA-protein interactions with LlamaTags—that together constitute a “microscope” for visualizing and interrogating the activity of genetic circuits as they function. We envision that LlamaTags can be applied to quantitatively measure the flow of information along regulatory networks in any multicellular organism that is amenable to transgenic control and live imaging. LlamaTags literally light up the central dogma, in real time, as development unfolds. Importantly, we can do more than map input to output: we can quantitate the dynamics of these connections over time. That’s the difference between trying to create a stop-motion movie where you need a new actor for each frame (a dead embryo), and actually observing development unfold in real time.
Physical Biology of Living Embryos
Like many of you, we believe that in order to construct a system, we should first understand it in quantitative detail. We view this as a call for reaching a “predictive understanding” of development, through which we can calculate developmental outcomes from knowledge of the concentrations of input transcription factors and the DNA regulatory sequence. To reach this predictive understanding, we believe that a powerful dialogue is necessary in which theoretical models make predictions that are subsequently tested experimentally, with measurements fed forward into the model to generate a new cycle of experiments. Our work in Cell is a crucial early step toward enabling the biophysical dissection of developmental programs by making it possible to measure the very same input-output functions predicted by our theoretical models.


 (6 votes)
(6 votes)