Viewing less to see more
Posted by Hiroki Ueda, on 23 October 2015
Dimitri Perrin3, Shimpei I. Kubota1,2, Kazuki Tainaka1,2 & Hiroki R. Ueda1,2,4*
1Department of Systems Pharmacology, The University of Tokyo, Tokyo, Japan.
2CREST, Japan Science and Technology Agency, Saitama, Japan.
3School of Electrical Engineering and Computer Science, Science and Engineering Faculty, Queensland University of Technology, Brisbane, Australia.
4Laboratory for Synthetic Biology, RIKEN Quantitative Biology Center, Osaka, Japan.
Correspondence should be addressed to H.R.U. (uedah-tky@umin.ac.jp, Tel: +81-3-5841-3415, Fax: +81-3-5841-3418)
The brain is an organ like no other, in part because of its function. It has been recognised as the location of imagination, memory, thought and sensation since Claudius Galenus, but details about its structure only start to emerge in the late 17th century. Thomas Willis proposes the concept of a regionalisation of brain activities and Antonie van Leeuwenhoek’s work on microscopy reveals that nerves are not hollow conduits for ‘animal spirits’.
While further advances (such as Luigi Galvani on the role of electricity in the nervous system) move 18th century scientists closer to understanding the brain, the mystic surrounding the organ remains after Matthias Jakob Schleiden and Theodor Schwann propose their cell theory. All organs follow the three tenets that all living organisms are composed of one or more cells, the cell is the most basic unit of life, and all cells arise from pre-existing, living cells. All organs, except the brain. Dyes used to reveal the cellular structures of tissues only show a dense and entangled network of fibres. No cells are visible in the brain.
Fifty years later, Santiago Ramón y Cajal starts using Camillo Golgi’s reazione nera, which has the distinct advantage of staining a limited number of cells at random and in their entirety. Fine details can, finally, be observed. Ramón y Cajal pushes forward the idea of a modular brain and cells as emitter/receptor. By viewing less, the method allows to see more.
While EEG, implants and fMRI now allow measurements of group cells or indirect observation of overall activity patterns, light diffraction due to lipids means that brains cannot be directly imaged. Slicing is still used, and requires time-consuming and error-prone computational reconstruction of the whole organ.
Tissue clearing, by contrast, removes these lipids and finally allows high-resolution whole-brain imaging, therefore preserving important structures. It is a crucial step, and it is fitting that Karl Deisseroth and his team show a transparent brain sitting over a Ramón y Cajal quote in their 2013 article on CLARITY. By viewing less, we can see more, but seeing is only the beginning.
Learning, memory, behaviour and all other cognitive functions emerge from structure and cell-to-cell interactions, making understanding cellular circuits in the brain essential to advances in Neuroscience. Coupled with key technologies such CRISPR/Cas-mediated genetic engineering, tissue clearing has the potential to have for this field the impact that microarrays had on Genetics.
This requires two properties: (i) tissue clearing must be safe, rapid, efficient and easily reproducible, (ii) computational tools must be developed to analyse these new high-resolution 3D images. Our method, CUBIC, has been developed to address these needs.
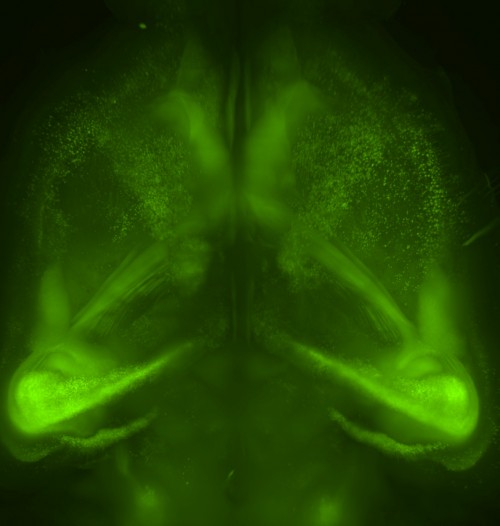
Our new aminoalcohol-based clearing cocktails have no safety concerns and preserve signals from fluorescence proteins. Whole-brain clearing can be achieved by immersing a whole brain in CUBIC reagents for two weeks. In our 2014 Cell article, we showed this protocol is also applicable to a marmoset brain, which is a model animal closer to the human.
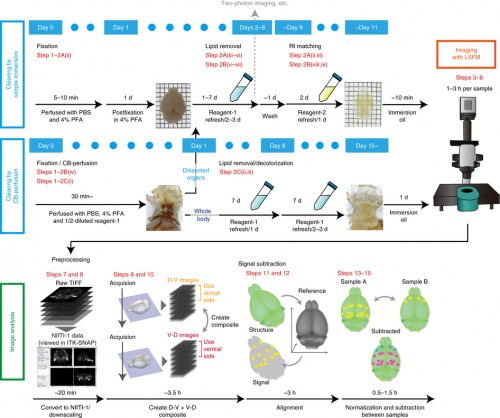
For the biologist end-user, imaging the cleared sample is a largely automated process. Our vision is to make the analysis as straightforward, with information about the experimental setup enough to identify the anatomical brain regions where changes in expression occur (and estimate the statistical significance of these changes). We have already developed tools for these analysis steps, with an initial pipeline described in our recent Nature Protocols article and available for download, and we are now working on improving the registration steps and the detection of active cells.
We have also shown that tissue clearing is useful not only for the brain but also for other organs and for whole-body imaging. Because we noticed aminoalcohols in the CUBIC reagent could decolourise the endogenous pigments such as heme, we developed a direct transcardial perfusion of the CUBIC reagent for further transparency. This perfusion protocol enables whole-body or whole-organ clearing within 10 days to 2 weeks.
Our new protocol provides access to a new world. CUBIC makes it possible to visualise and quantify a targeted small minority cells in the 30 billion cells of a mouse body. This is helpful to understand cellular mechanisms of autoimmune disease and cancer micrometastasis. Of course, our new protocol is not limited to model organisms expressing fluorescent proteins. CUBIC is compatible with immunohistochemistry so we can apply our method to human pathology, for which fluorescence imaging is not possible.
Further reading:
- K. Chung, J. Wallace, S.-Y. Kim, S. Kalyanasundaram, A. S. Andalman, T. J. Davidson, J. J. Mirzabekov, K. A. Zalocusky, J. Mattis, A. K. Denisin, S. Pak, H. Bernstein, C. Ramakrishnan, L. Grosenick, V. Gradinaru and K. Deisseroth (2013). Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. DOI: http://dx.doi.org/10.1038/nature12107
- E. A. Susaki, K. Tainaka, D. Perrin, F. Kishino, T. Tawara, T. M. Watanabe, C. Yokoyama, H. Onoe, M. Eguchi, S. Yamaguchi, T. Abe, H. Kiyonari, Y. Shimizu, A. Miyawaki, H. Yokota and H. R. Ueda (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739. DOI: http://dx.doi.org/10.1016/j.cell.2014.03.042
- K. Tainaka, S. I. Kubota, T. Q. Suyama, E. A. Susaki, D. Perrin, M. Ukai-Tadenuma, H. Ukai and H. R. Ueda (2014). Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924. DOI: http://dx.doi.org/10.1016/j.cell.2014.10.034
- E. A. Susaki, K. Tainaka, D. Perrin, H. Yukinaga, A. Kuno and H. R. Ueda (2015). Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging with single-cell resolution. Nature Protocols 10, 1709–1727. DOI: http://dx.doi.org/10.1038/nprot.2015.085


 (4 votes)
(4 votes)