“My balance comes from instability” thought Herzog (Saul Bellow, Herzog)
Posted by zelhanil, on 28 October 2019
A bit of background
The dependence of a protein’s function on its structure is a well-known phenomenon. Back in 1970’s, it was suggested that most proteins would fold into one energetically stable or favorable conformational state in the cell determined by their primary amino acid sequence. This led to the notion of “one sequence to one structure to one function”. Later, the identification of prions which have more than one stable structure or conformation in the same cell suggested there are exceptions to this rule. However, in the early days of this discovery, one of these conformations was nonfunctional and disease causing; therefore, the dogma still held true. Later, the discovery of functional non-toxic counterparts of prions, called prion-like proteins, challenged the dogma. These proteins not only existed in different conformations but also had different functions associated with different conformations, expanding the functional space proteins can occupy.
Today, prion-like proteins are shown to have roles in different physiological processes including adaptation to changing environmental conditions, immune response and memory formation. In all of these processes, they act as transcriptional or translational regulators, or signaling components at the molecular level, leading to a global change in cellular response. Most of these prion-like proteins are well-studied yeast prions and are often determinants of heritable phenotypes. When we look at higher order organisms, the examples of such proteins are restricted. Recent computer-based screens show these proteins are prevalent throughout all kingdoms of life; yet it is still unknown what functions they serve in different conformational states in normal physiology of higher order organisms.
Walking on Drosophila Proteome
Kausik Si’s research lab (https://research.stowers.org/silab/) at Stowers Institute for Medical Research in Kansas City is home to the very first prion-like protein found in Drosophila, called Orb2, which is important for the persistence of memory. The discovery of Orb2 and the commonality of prion-like proteins in yeast led to the idea that prion-like proteins could work in many other physiological processes. The Si lab conducted a small-scale computational screen on the Drosophila proteome to find out novel prion-like proteins. Focusing on the top scoring proteins, his team performed an experimental screen using exogenously expressed versions of the selected candidate proteins with various conventional techniques used to characterize prion-like properties (see the publication for technical details). This eventually led to a small set of 5 proteins that could be used to explore the effects of their conformation on their functions in vivo. Some of these proteins have well-known functions, and the only thing needed was to relate the conformations to these functions. The most challenging part was analyzing the proteins which did not have any functional information but only a protein sequence. Herzog, once called CG5830, was one such protein. It was at this point as a naïve but venturesome graduate student, I came into the picture!
Things got a little bit spicier
What one does when faced with a newly discovered completely unknown protein: a homology search to find out what similar proteins do in other organisms. Herzog’s homolog in mammals is a phosphatase, called SCP1, which regulates RNA polymerase II activity. When I took over the Herzog project, I first did several experiments to understand whether Herzog is really a homolog of SCP1. However, I found that even though Herzog has in vitro phosphatase activity, it doesn’t have functional similarity to SCP1 in the cell. So, I was back to where I started.
A protein has different roles at each level of biological organization, from molecules to organism, and the particular functions it performs at one level add up and lead the way to the next level, eventually telling us how that protein contributes to the life of the organism. Now, to understand Herzog completely, I needed to systematically characterize its functions at the molecular, cellular, and organismal levels. I had a lot of questions: Is it a phosphatase in the cell? If so, what are the substrates? Where does it localize? What does it partner with to do its job? What does it control? How important it is for the fly? Finally and most importantly what is the connection between its conformation and function? To answer all these questions, I needed a flexible medium of work. Luckily enough, I was working with Drosophila melanogaster, which has various tools and resources making it easy to manipulate at all these different levels.
Herzog all the way down
Before I get into the details of our story, I want to point you to the illustration below to put things in context. Along with our publication, I decided to submit potential cover images. One of those images was drawn by Stephanie Nowatarski, PhD (a really good old Drosophilist, now Planari-st and artist; http://stephanienowotarski.com/) in collaboration with Mol Mir (a researcher and art-maker; https://molmir.com/). It depicts our model from bottom (molecular structure) to top (animal viability)- highlighting the scalar structure of the analysis we performed in the paper.
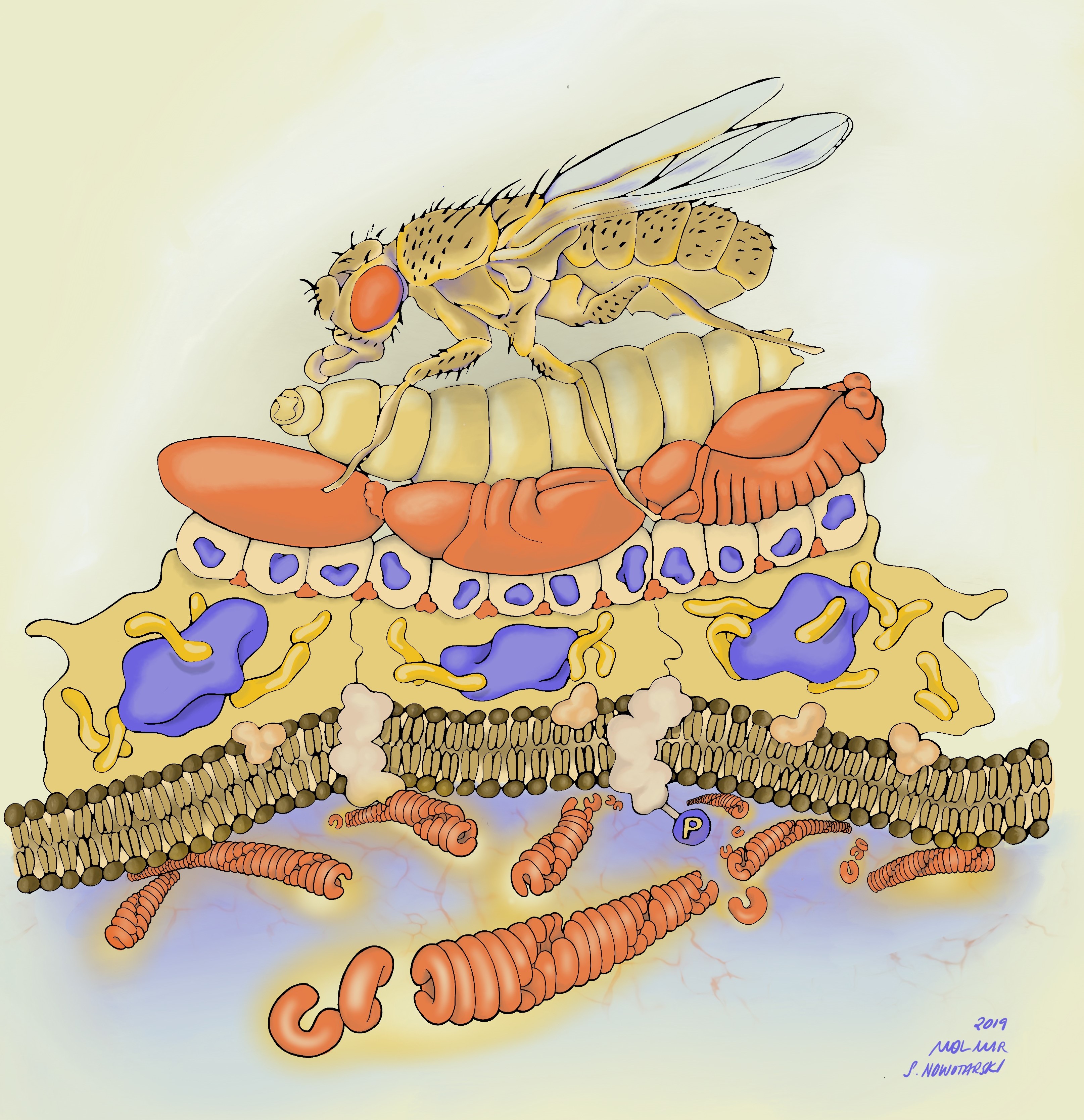
The two main strategies I followed to discover the function of Herzog were: 1- To look for the phenotypic effect of herzog mutations on the organism; and 2-To follow its expression pattern over time and space.
CRISPR-derived herzog mutant lines, which lack the middle phosphatase domain and the rest of the C-terminal of the protein, were embryonic lethal in the F2 generation. To understand when and where they had defects during embryonic development, I first examined their cuticles. I observed that mutants do not have the characteristic denticle pattern of a differentiated embryo. Referring to the famous “Heidelberg screen”, it was clear that herzog mutant embryos had defects in segment polarity, which is regulated mainly by two genes, i.e., engrailed and wingless. In embryos, engrailed starts expressing just after cellularization at the beginning of gastrulation. Staining of herzog mutant embryos right at this stage for engrailed showed us a diffused expression of engrailed in these mutants compared to wild type. When I looked at wingless, which is normally expressed at the posterior end of each segment later in development, I observed that while herzog mutants had the correct number of segments, there was an additional stripe of wingless in each segment (this data did not make it to the paper). This suggested that the diffused expression pattern of engrailed led additional cells to gain wingless identity, causing segments to lose their polarity. Most importantly, this told us that herzog has a role in establishing the A-P axis of each segment, acting as an antagonist of wingless signaling which begins with gastrulation. But how about the normal expression pattern of herzog? For that, the herzog mutant phenotype suggested two possibilities: 1- Herzog protein is both expressed and functional at this specific time point, like the other patterning genes, or 2-Herzog is there all along, but something happens at this time point such that the protein becomes functional.
Let’s find the answer! For that, I endogenously tagged Herzog and followed it in the fixed embryos. I observed that Herzog is ubiquitously expressed on the membrane throughout embryonic development. In contrast to the patterning genes it regulates, Herzog does not have a specific expression pattern, which ruled out the first possibility I suggested above. Then I started thinking how I go about the second possibility. Wait a minute! I had embryos with fluorescent Herzog which I could follow live and focus closely! Collaborating with an awesome microscopist, @jeff_j_lange, we saw something happening to the protein just at the start of gastrulation: it is diffusely localized along the membrane until the end of cellularization, and when gastrulation begins, it changes to a punctate form on the membrane, and the punctate pattern persists throughout the rest of embryonic development. Now, I had a phenotype correlating a physical state change in the protein, which raised the curious question: Are the conformations of these two different physical states of Herzog different as would be the case for a prion-like protein? Before answering this question, I should mention that prion-like proteins can exist in at least two different conformations in the same cell: a monomer and an aggregate which can attain various physical conformations with different stabilities, ranging from flexible liquid droplets to very stable amyloids. To understand Herzog’s conformations, I followed it by western blot analysis at different stages of embryogenesis, and I saw that it exists as soluble low molecular weight monomers in all stages of embryogenesis. Interestingly, it transforms into high molecular weight aggregates, just after gastrulation, which correlates with the timing of its punctate pattern. Insolubility of these aggregates, even under the extreme denaturing conditions of boiling and detergents, suggests a stable conformation, like amyloid. In line with this idea, I found that Herzog protein staining colocalizes with an amyloid specific dye in embryos and that purified embryonic Herzog protein can be recognized with amyloid specific antibodies. So, I now had a protein that changes to an amyloid like state during gastrulation, and this coincides with its segment polarity phenotype. However, I still needed to know what this means at the molecular/biochemical level.
To understand the molecular function of Herzog, I thought I could build a reporter system to detect Herzog’s enzymatic activity in the presence or absence of aggregation. For this, I needed to answer two important questions: 1- Which part of the protein causes it to aggregate? and 2- What are the substrates of Herzog? Using truncated versions of Herzog, I found that N terminal prion-like domain of Herzog is responsible for both its aggregation and membrane localization. With proteomic analyses, I found that it interacts with well-known developmental regulators, which have roles in TGFβ/BMP, EGF and FGF signaling pathways and cell cycle. Focusing on one potential candidate called Dah, which was previously found to be dephosphorylated during gastrulation by an unknown phosphatase, I designed an enzymatic assay for Herzog and found that Herzog dephosphorylates Dah, for which N terminal prion-like domain is required. Replacing Herzog’s prion-like domain with a known amyloid forming prion-like domain (which also had a membrane targeting motif) rescued the enzymatic activity. This meant that the phosphatase activity of Herzog depends on its amyloid-like aggregation through its N terminal prion-like domain on the membrane. However, this did not tell us whether there is a clear distinction between activities of monomers and aggregates. In collaboration with an experienced structural biologist, Ruben Hervas Milan, we recapitulated the enzymatic activity of Herzog aggregates with purified protein from embryos. Importantly, we found that aggregates were active, and monomers did not have phosphatase activity. Moreover, when we allowed monomeric recombinant Herzog protein to self-assemble into amyloid-like fibrils, we observed a dramatic increase in enzymatic activity, while dissociation of these fibrils with an amyloid inhibitor abolished the activity.
Putting all these pieces together, I can now say that Herzog’s switch into amyloid-like aggregates is a developmentally regulated process resulting in its enzymatic activation that is essential for the patterning of the embryo.
What is ahead of us?
Although it was previously shown that knock down of prion-like homolog of prion protein in PrP knock out mouse results in an embryonic phenotype, how or whether the conformational change of these proteins affects development has not been studied. Herzog exemplifies for the first time how a protein conformational switch into a higher order amyloid state regulates a specific process in embryonic development. As the first natural example of an amyloid enzyme, Herzog demonstrates an alternative mode of enzymatic regulation: the use of a prion-like domain to regulate catalytic activity with conformational change. Looking ahead, we want to understand how the amyloid structure orients the catalytic domain of the enzyme to alter its activity; how the conformational switch of Herzog is regulated during development and how the aggregation dynamics regulate embryonic patterning.
Looking at its interactors, Herzog seems to have several potential functions at the intersection of multiple developmental signaling pathways. These pathways are inherently dynamic but lead to stable information to carry development forward. How can an enzyme with a seemingly stable conformation like amyloid make it in the dynamic environment of signaling pathways and so in development? Recent studies have shown that identical polypeptides can fold into multiple, distinct amyloid conformations and that amyloid structure can dynamically form and disappear via post translational modifications. We speculate that such structural flexibility and heterogeneity would allow a protein like Herzog to adopt stable yet dynamic conformational states. Moreover, these features may also lead to functional diversity such that a an amyloid-like protein can form distinct functional units, with even opposing functions, in the same cell and can maintain this functional diversity with the help of its stability. Development, which needs to accommodate the changing environment, might utilize such molecular stability and flexibility to tune the time course of development. Therefore, our study lends support to the idea that there can be other prion-like conformational functional switches regulating other important developmental events.
Read the original article:
Amyloid-like Assembly Activates a Phosphatase in the Developing Drosophila Embryo
Nil, Z., Hervás, R., Gerbich, T., Leal, P., Yu, Z., Saraf, A., Sardiu, M., Lange, J.J., Yi, K., Unruh, J., Slaughter, B., Si, K.
Cell 2019. DOI: 10.1016/j.cell.2019.08.019
A highlight by Paulina Strzyz:
Amyloid-directed phosphatase activation
Nat. Rev. Mol. Cell Biol. 2019. DOI: 10.1038/s41580-019-0175-6


 (6 votes)
(6 votes)