A tale of two fishes
Posted by Krista Gert, on 25 August 2023
Krista Gert, a recent doctoral graduate from Andrea (Andi) Pauli’s lab at the Research Institute of Molecular Pathology (IMP) in Vienna, Austria, recently published a study on how Bouncer, a small egg membrane protein required for sperm binding, governs compatibility between the sperm and eggs of different fish species. Her study, published in Nature Communications in June, juxtaposes two distantly related, reproductively isolated fish species—zebrafish and medaka. Using a structure-function approach guided by evolutionary analysis, her work uncovers what makes a Bouncer protein compatible with one species’ sperm but not another’s, providing an intriguing molecular explanation for fish hybridization. We caught up with Krista to find out more about the story behind the paper.
What brought you to Andi’s lab and how did the project start?


I first heard about Andi’s lab through an advertisement for the Vienna BioCenter Summer School program back in 2016 while I was doing my master’s degree at Linköping University in Sweden. I had previously participated in a summer REU (Research Experience for Undergraduates) program while a bachelor’s student and was keen on doing a summer research project again. Developmental biology fascinated me, so I applied to Andi’s lab because of a project on a small protein called Toddler and its role in regulating cell migration during gastrulation. I enjoyed being in Andi’s lab and working with zebrafish that summer so much that I came back for my master’s thesis project in 2017. For that project, I changed gears from working on Toddler to Bouncer and its role in fertilization with a then Ph.D. student, Sarah Herberg. The work I did during that time set the stage for my own Ph.D. project and led to my discovery that Bouncer keeps fertilization species-specific between medaka and zebrafish—a major finding in our first publication on Bouncer in Science. These exciting results provided the momentum for my own Ph.D. project. I dove into exploring how Bouncer mediates specificity in sperm-egg interaction between species, the main goal of my paper in Nature Communications.
What was known about cross-fertilisation between different species before your work?
In general, scientists had observed a high frequency of hybridization among fish species compared to other animal groups, but no one had really sought to understand the molecular basis for it given the fact that we didn’t even know what proteins are generally required for sperm-egg membrane interaction in fish. That’s part of why it was so exciting to work on Bouncer—it was the first egg molecule shown to be required for fertilization in any fish species, opening the door to not only studying its function in sperm binding, but also probing how it might limit binding to only sperm from closely related species or allow hybridization across greater phylogenetic distances.
Can you summarise your key findings?
When I started working on Bouncer, I initially observed that Bouncer enabled fertilization in a species-specific manner: eggs expressing zebrafish Bouncer could be fertilized by zebrafish sperm, but not by medaka sperm. At the beginning of my current study, however, I soon found that there was more to it than that—zebrafish sperm turned out to be compatible with several different species’ Bouncer proteins, and remarkably, some Bouncer proteins could even work with both medaka and zebrafish sperm.
However, taking advantage of Bouncer’s medaka/zebrafish specificity, I defined features within Bouncer orthologs, both on the amino acid and post-translational modification levels, that are required for interaction with zebrafish or medaka sperm. In terms of evolution, we found that Bouncer remains largely similar from fish species to fish species, yet we identified a medaka-specific change in the amino acid sequence (site 63) which contributes to the medaka/zebrafish cross-fertility block. In zebrafish, this site contains arginine (R), whereas medaka Bouncer contains a leucine (L). We furthermore found that the N-glycosylation pattern of Bouncer really matters for medaka sperm compatibility but does not influence zebrafish sperm compatibility. In this way, I elucidated some of the molecular details as to why zebrafish and medaka cannot cross-fertilize and furthermore demonstrated that there are different levels of stringency for sperm-Bouncer interaction depending on the species, likely impacting their ability to hybridize with more distantly related species vs. only close relatives.
Were you surprised to find that seahorse and fugu Bouncer are compatible with both zebrafish and medaka sperm?
Yes, completely! This result was particularly surprising when considering that both seahorse and fugu Bouncer are ~40% identical to zebrafish Bouncer, whereas medaka Bouncer shares just under 39% identity with zebrafish Bouncer. As I mentioned before, it was also quite a surprise since we originally expected that Bouncer would show strict species specificity beyond just the medaka/zebrafish combination. Nonetheless, it was an informative result because I could then focus on sequence elements that were different between medaka and zebrafish Bouncer and ignore any changes that were also present in fugu and seahorse Bouncer, allowing me to narrow the field in determining what underlies Bouncer’s medaka/zebrafish specificity.
What was it like working with two different species of fish?

For me, working with animals is one of the most enjoyable parts of being a biologist, so I gladly accepted the challenge of introducing a new model species into the lab. Before my project, we had only zebrafish, and I needed to set up all the protocols and tools for working with medaka. It was not easy, but I found it very rewarding once I got everything up and running! Having the side-by-side comparison of medaka and zebrafish was also very informative in terms of their intrinsic species differences, particularly in the process of fertilization itself.
Did you have any particular result or eureka moment that has stuck with you?
My biggest “eureka moment” would have to be when I saw zebrafish eggs fertilized by medaka sperm for the first time. During my master’s project, I had made several bouncer mutant zebrafish lines expressing other species’ bouncer genes to test whether they could rescue fertilization with zebrafish sperm. I tested the Bouncer proteins from human, mouse, Xenopus, and medaka, but none of them were compatible with zebrafish sperm. But what about the other way around? Could I fertilize these zebrafish eggs with sperm that was species-matched with the expressed Bouncer protein? The scenario most likely to work was medaka sperm with medaka Bouncer-expressing zebrafish eggs, so I procured several medaka males from a neighboring lab and gave IVF a try. After a few attempts and to my great surprise, I caught sight of cleavage-stage embryos in the petri dish as I peered through the dissection scope a few hours later—the very first zebrafish-medaka hybrids to ever exist!
And the flipside: were there any moments of frustration or despair?
When I started this project, I believed that I would be able to pinpoint the exact changes needed to make zebrafish Bouncer compatible with medaka sperm. Each time I made a new line, I thought for sure that this would be the one that would finally work. There is a significant time lag between making a new line and getting the results since you need to wait for two generations, and it takes about 3 months for zebrafish to reach sexual maturity. After making and testing all my amino acid substitution mutants without much luck, I was convinced that combining the R63L change with the medaka N-glycosylation pattern in zebrafish Bouncer would be sufficient for medaka sperm to work. It was disappointing when it didn’t, and even more so when I then tried the entire medaka finger 3 sequence plus the N-glycosylation pattern in zebrafish Bouncer, and this still failed to work with medaka sperm. Looking back, though, I am glad I kept trying even though I didn’t quite solve the entire puzzle of species specificity.
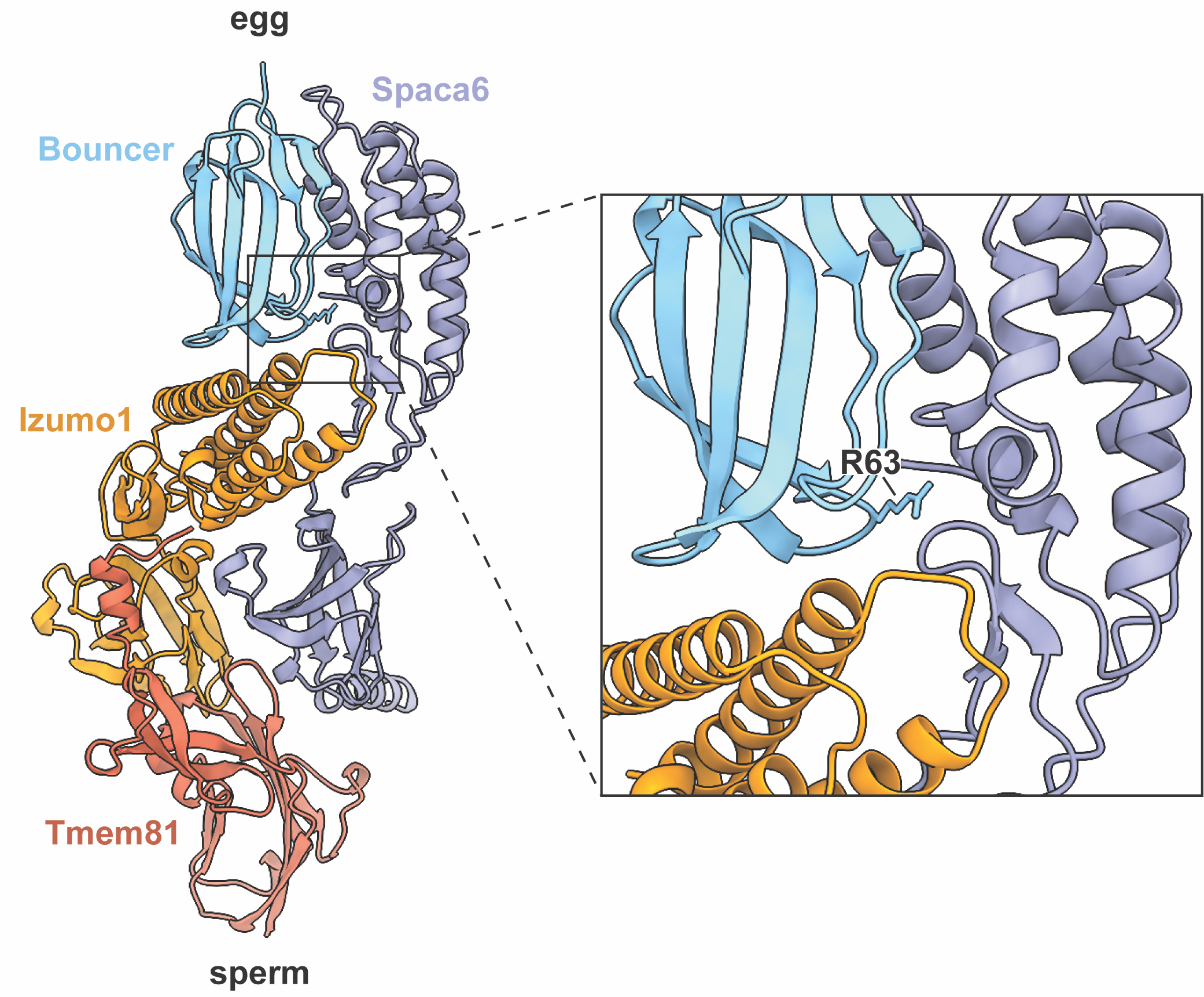
My initial discovery of Bouncer as the main block to cross-fertilization between medaka and zebrafish provided a versatile system for uncovering Bouncer’s interaction partner on sperm. On top of this, my current paper highlights the importance of site 63 in determining specificity, and it may therefore also be critical for interaction with the sperm binding partner. Current work in the lab by my colleagues Andreas Blaha and Victoria Deneke uncovered that Bouncer may interact with a heterotrimeric complex on sperm consisting of Izumo1, Spaca6, and a new factor, Tmem81, as they report in their recent bioRxiv preprint (Deneke, Blaha, et al., 2023). This is groundbreaking for the fertilization field since the only sperm-egg protein interaction pair of which we know currently is IZUMO1 and JUNO in mammals. What’s especially striking to me is that when you look at the AlphaFold-predicted model of zebrafish Bouncer and the sperm trimer, the tip of finger 3 where R63 sits in Bouncer is right at the interface between Spaca6 and Izumo1. It’s exciting to see how my work was able to pinpoint a potential key interaction site even though I had only half of the picture. Now that we know Bouncer’s interaction partner on sperm, we can work toward defining the Bouncer-trimer interaction interface and understand why zebrafish sperm are able to bind many Bouncer orthologs, whereas medaka sperm maintain higher specificity.
And personally, what is next for you after this paper?
Discovering how to make hybrids between medaka and zebrafish spawned another whole project on which I embarked during my Ph.D. I used them to address another question of species specificity: how is the timing of zygotic genome activation (ZGA) determined? Medaka and zebrafish have proved to be a very interesting comparative system for this study as well, given their different times of ZGA onset during embryogenesis. Currently, I’m finishing up work in the lab for a paper on how we used these hybrids to explore this question. In case you are curious, there’s already a preprint on bioRxiv (Gert et al., 2021).
I defended my Ph.D. earlier this summer and am currently looking for a post-doc position to continue working on fertilization or reproductive biology. I am especially excited to work on understudied species and to establish new animal models, as my work with Bouncer in two distantly related fish species has really convinced me of the value of a comparative system. In the future, I hope to work with endangered species and develop new reproductive technologies to aid their conservation.


 (1 votes)
(1 votes)