An interview with Marco Podobnik, GfE PhD award winner 2024
Posted by the Node, on 25 April 2024

The 2024 German Society for Developmental Biology (GfE) PhD award recipient was Marco Podobnik, who worked on pigment pattern diversification in Danio fish species at the Nüsslein-Volhard laboratory in Tübingen. We caught up with Marco to learn more about his background, his PhD work and his research interests as a postdoc at the Australian Regenerative Medicine Institute.
First of all, congratulations on receiving the 2024 GfE PhD award! What does this award mean to you?
Thank you. I am only half-way joking when the first thing that comes to my mind is, hey, now I can fill out the line that asks for “prizes” in grant applications. Being recognized by the German Society for Developmental Biology is a huge honour. The fact that my work was so collaborative makes it important to draw the attention to my colleagues I worked with over the years. The Max Planck Society creates permissive environments for basic research we conducted, a privilege I am aware of. I wish these opportunities would be more accessible.
Let’s go back to the beginning. When did you first become interested in science?
I believe the way we construct stories of the scientific process is a symptom of the humane desire to tell good stories. This also applies to the story of people’s life history. When I was a kid, I always liked to be out in the forests, trying to identify birds by their sounds. Although I was never really excited about maths, I deliberately chose all science subjects in high school. I had an excellent chemistry teacher, Holger Mummert, who did his PhD in Tübingen. In my final year I joined his class on fossils which I found beautiful. I had to become a biologist, right?
How did you come to do a PhD at the Nüsslein-Volhard laboratory?
I think this destination was the consequence of a random process paired with a developing passion. When I still studied biology at the University of Cologne, I had to find a lab for an internship. So I made a list with shiny labs working on topics I mostly never heard of and brought it to my mentor, Matthias Hammerschmidt. He had done his PhD with Christiane (Janni) Nüsslein-Volhard in Tübingen. In the early 1990s, he participated in the genetic screens in zebrafish [1], a heroic effort approaching the Nobel Prize work in flies in the 1980s [2]. When I showed Matthias the list, he smiled and refused to send me anywhere. Instead, he offered me to take a look into his zebrafish lab. Seeing live zebrafish embryos under a stereomicroscope for the first time was quite a fascinating moment. After that he suggested me to go to the Max Planck Institute for Developmental Biology in Tübingen (now the MPI for Biology).The real deal was the weekly meetings in Janni’s small office, where everybody had to hunch shoulders to fit in. Tiny zebrafish summits regularly joined by Patrick Müller who led a group at the Friedrich Miescher Laboratory next door.
Back in Cologne I obtained my undergraduate degree with Sigrun Korsching, who had been a junior group leader in Tübingen before she established her zebrafish lab in Cologne. At some point Uwe Irion offered me a PhD position in Janni’s lab. I had just come back from two expeditions with other students from the Cologne University on microbial communities in South America and in the North Atlantic Ocean. Biodiversity and development were (and still are) exciting to me, so I chose to work with Uwe on pigment patterning in other Danio species originally coming from Southeast Asia.
Can you summarise your PhD research?
Although Janni’s lab had always been interested in Danio species other than the zebrafish [3, 4], the pioneering work came from David Parichy and colleagues over the last two decades [5, 6]. A milestone was the release of a reliable phylogeny of the genus by Braedan McCluskey and John Postlethwait [7]. I found it fascinating that species most similar in their pigment patterns were not necessarily most closely related, and most closely related species often had most divergent patterns. Zebrafish develop a striking pattern of horizontal blue and golden stripes, while the sister species D. aesculapii conceals itself by forming dark vertical bars that are low in contrast. Given the knowledge about the genetic basis of stripe formation in zebrafish, maybe we could identify the genes that functionally diverged to contribute to patterning differences between species.
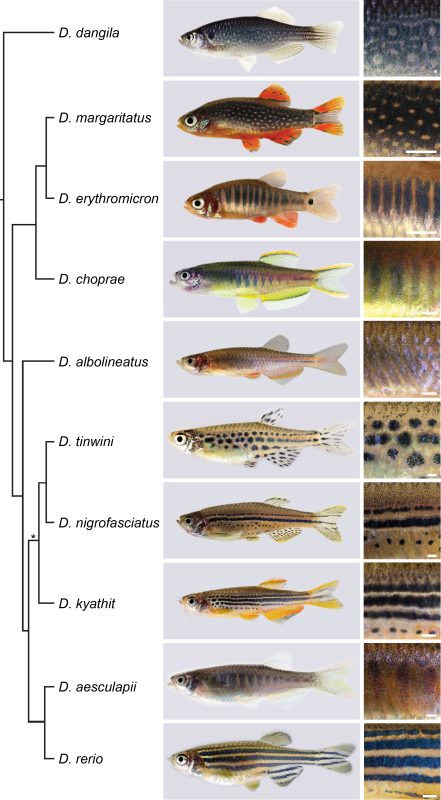
We decided to focus on genes that code for proteins mediating direct cell-cell contacts. Uwe had already generated mutants in a handful of these genes and acquired a number of Danio species; a collection I helped to make more comprehensive. We were inspired by the reciprocal hemizygosity test to identify diverged genes and we could meet the requirements to apply it, namely generating null mutants in species and hybrids between species [8]. Such an endeavour in non-model organisms was unthinkable without the CRISPR/Cas9 system, which the lab aims to improve since its adaptation for genetic engineering [9, 10]. Uwe and I started an extensive effort of making hybrids, pairing wild-type and the newly generated mutant species in various combinations. Four years later it all came together: While some genes remained functionally conserved, others diverged across the phylogeny [11].
In the case of the potassium channel gene kcnj13 we identified a repeated and independent functional divergence; this contributed to patterning differences between zebrafish and D. aesculapii [12]. We found that the black melanophores require the kcnj13 function to acquire certain cell shapes within their own population but they also instruct the two other pigment cell types, yellow/orange xanthophores and shiny/blue iridophores, to change their shapes according to their location in the skin. As the D. aesculapii allele is functionally different from the zebrafish one, we propose that divergence in kcnj13 caused changes in the way the pigment cells interact and change their shapes to contribute to the species-specific differences in colour and contrast of the patterns [13].
Your PhD research involves working with a wide range of techniques. Do you have a favourite?
I love genetics for its power to let us understand processes across different levels of biological organisation. During university I learnt about Muller’s classification of alleles [14]. It was beautiful to see how it could be used when we studied pattern development in hybrids between Danio species. The altered patterns in hemizygous hybrids between mutant zebrafish and wild-type D. aesculapii indicated that the kcnj13 allele from D. aesculapii behaves like a hypomorph as it could not compensate the CRISPR/Cas9-induced loss-of-function of the D. rerio allele. The patterns of hemizygous hybrids from the reciprocal cross are similar to the patterns of hybrids between wild-type species. Thus, the wild-type alleles from zebrafish and D. aesculapii cannot be functionally equivalent. Initially we thought that the D. aesculapii allele had lost its function completely but mutant D. aesculapii indicated that the gene function was still required for patterning. I love discovering obvious phenotypes in mutants, as it can be a rare but very profound experience. The suitability of zebrafish for fluorescence imaging in vivo makes it a fantastic model, as you can see cells behaving in real time, sometimes even with subcellular resolution. I can probably make the most meaningful contributions by applying a duet of genetics and in vivo imaging. Often the successful outcome of any experiment relied on the groundwork and expertise of my colleagues. I essentially understand science as teamwork.
Speaking of teamwork, how was your experience collaborating with people across the world for your PhD work?
As you can imagine pigment patterning is a relatively small field, although it hopefully becomes apparent how exciting it is. It’s great that people with expertise in genetics, sequencing methods, protein structure modelling and image analysis collaborate on this topic. I feel there aren’t really boundaries when it comes to common interests and curiosity, it’s just essential to bring passionate people together. It’s important to communicate effectively and to make it fair for the people involved. When it comes to generating ideas, bigger meetings might not always be effective. The most creative ideas emerged during our small lab and one-to-one meetings.
Were there any frustrating times during your PhD? And on the flip side, any particularly memorable moments?
My most exciting moments were the ones when we made discoveries in the lab. Sometimes it took a while to realize them. It requires one to think about observations over and over again. We were all working a lot, basically every day including most weekends, which is an intensity I have decided to reduce. I have fond memories of the garden parties in summer and the yearly Christmas cookie baking at Janni’s house.
You’ve recently moved across the globe to do a postdoc in Melbourne. How was the experience and what motivates your research today?
The Melbourne metropolitan area is a great place for personal life and research, although it can be depressing to think about the fate of the traditional owners of this country. The Australian Regenerative Medicine Institute cultivates broad interests in development, regeneration, evolution and medicine. Our lab headed by Peter Currie explores muscle biology using a spectacular diversity of fish models. It’s stimulating to be part of a bigger team. I find it exciting to help others with their projects, while I am defining my own long-term goals.
Finally, let’s go outside of the lab. What do you like to do in your spare time?
I play the saxophone in various settings for over 20 years now, which is tremendously important to me. I also love hiking. A very memorable trip was the Via Alpina in Switzerland and now very recently a trip to the Victorian Alps in Australia.

Dr. Marco Podobnik
Research Fellow
Australian Regenerative Medicine Institute, Monash University, Clayton 3800 VIC, Australia
X: @m_podobnik
Website: https://marcopodobnik.wordpress.com/
References
1. Nusslein-Volhard, C. (2012). The zebrafish issue of Development. Development 139, 4099-4103.
2. Wieschaus, E., and Nusslein-Volhard, C. (2016). The Heidelberg Screen for Pattern Mutants of Drosophila: A Personal Account. Annu Rev Cell Dev Biol 32, 1-46.
3. Singh, A.P., and Nusslein-Volhard, C. (2015). Zebrafish stripes as a model for vertebrate colour pattern formation. Curr Biol 25, R81-R92.
4. Irion, U., and Nusslein-Volhard, C. (2019). The identification of genes involved in the evolution of color patterns in fish. Curr Opin Genet Dev 57, 31-38.
5. Parichy, D.M., and Johnson, S.L. (2001). Zebrafish hybrids suggest genetic mechanisms for pigment pattern diversification in Danio. Dev Genes Evol 211, 319-328.
6. Patterson, L.B., and Parichy, D.M. (2019). Zebrafish Pigment Pattern Formation: Insights into the Development and Evolution of Adult Form. Annu Rev Genet 53, 505-530.
7. McCluskey, B.M., and Postlethwait, J.H. (2015). Phylogeny of zebrafish, a “model species,” within Danio, a “model genus”. Mol Biol Evol 32, 635-652.
8. Stern, D.L. (2014). Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet 30, 547-554.
9. Irion, U., Krauss, J., and Nusslein-Volhard, C. (2014). Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141, 4827-4830.
10. Dorner, L., Stratmann, B., Bader, L., Podobnik, M., and Irion, U. (2024). Efficient genome editing using modified Cas9 proteins in zebrafish. Biol Open 13.
11. Podobnik, M. (2023). On the Genetic Basis of Pigment Pattern Diversification in Danio Fish, (Eberhard Karls Universität Tübingen).
12. Podobnik, M., Frohnhofer, H.G., Dooley, C.M., Eskova, A., Nusslein-Volhard, C., and Irion, U. (2020). Evolution of the potassium channel gene Kcnj13 underlies colour pattern diversification in Danio fish. Nat Commun 11, 6230.
13. Podobnik, M., Singh, A.P., Fu, Z., Dooley, C.M., Frohnhofer, H.G., Firlej, M., Stednitz, S.J., Elhabashy, H., Weyand, S., Weir, J.R., et al. (2023). kcnj13 regulates pigment cell shapes in zebrafish and has diverged by cis-regulatory evolution between Danio species. Development 150.
14. Muller, H. (1932). Further studies on the nature and causes of gene mutations. Jones DF, ed. In Proceedings of the 6th International Congress of Genetics. pp. 213-255.


 (No Ratings Yet)
(No Ratings Yet)