Behind the paper story: uncovering non-canonical functions of the Hippo pathway
Posted by Heya Zhao, on 21 April 2023
Dr. Heya Zhao and Dr. Alexey Veraksa at the University of Massachusetts Boston and Dr. Kenneth Moberg at Emory University School of Medicine recently published an article in Developmental Cell where they reveal the non-canonical functions of the Hippo pathway in developmental cell fate decisions in the Drosophila eye. They find that the effect is exerted through the interaction between transcriptional coactivator Yorkie (Yki) and transcriptional intermediary factor 1/tripartite motif (TIF1/TRIM) family protein Bonus (Bon), as well as their regulation of a non-canonical transcriptional program.
1. How did you get started on this project?
When I started as a Ph.D. student in the Veraksa Lab, Alexey and Ken got a new grant on characterizing novel members of the Hippo pathway. We thought that identifying new components of the pathway could expand our understanding of the canonical function of the Hippo pathway in growth regulation and reveal the underlying molecular mechanisms. Given that the transcriptional activity is the ultimate output of the pathway, but the transcriptional machinery was not well understood at the time, we set out to identify binding partners of the transcriptional coactivator Yki through affinity purification-mass spectrometry. In the Yki protein interactome, we identified Bon as a novel interactor, which led us to the identification of completely different functions and mechanisms from what we set out to pursue.
2. Can you summarize your findings?
After identifying Bon in the Yki interactome, we found that Bon interacts with Yki through PPxY motifs in Bon and WW domains in Yki. Instead of regulating tissue growth and embryonic peripheral neuron specification, which represent the independent functions of Yki and Bon, the Yki-Bon complex promotes epidermal fate and antennal fate and suppresses the eye fate. We also studied the Bon interactome and found that the Yki-Bon complex regulates these cell fate decisions by recruiting multiple transcriptional and post-transcriptional regulators, including Sd, HDAC1, Su(var)2-10, Hrb27C, and Svb/Ovo. We then identified genes jointly regulated by Bon and Yki by doing transcriptome analysis, which revealed their repression of Notch targets (e.g. E(spl)-C) and activation of epidermal differentiation genes (e.g. sha, f). We further validated the unexpected repression of E(spl)-C in eye development using transcriptional reporters. This is the first evidence that Yki can function in transcriptional repression in a non-mammalian system. We then showed by genetic analysis that the Yki-Bon complex functions in eye-antenna-epidermis fate determination through their repression of E(spl)-C and activation of sha and f. Altogether, our work revealed a previously unappreciated function of the Hippo pathway in eye-antenna-epidermis fate determination. This function is exerted through the interaction between Yki and Bon, their recruitment of co-regulators, and the transcription of a unique set of target genes (Figure 1).
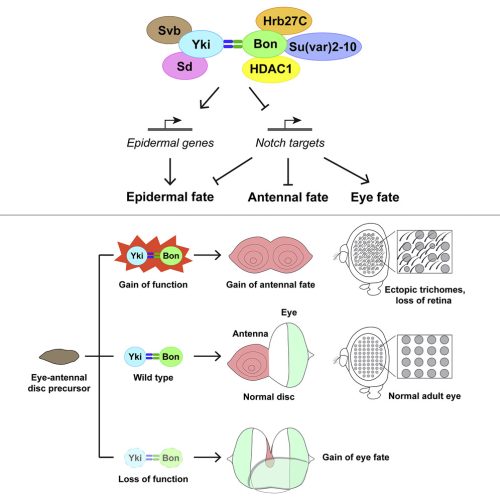
3. When doing the research, did you have any particular result or eureka moment that has stuck with you?
The first result that started to shape the paper was the rescue of Bon-induced epidermal hairs (trichomes) in the eye by knockdown of yki. We spent a long time trying to find a genetic interaction between Bon and Yki by studying their known phenotypes in tissue growth and embryonic peripheral neuron development, but this was unsuccessful. Then, while examining the growth phenotype in adult eyes, I noticed that the eyes look “hairy” under the dissecting microscope when Bon and Yki were overexpressed simultaneously. Upon closer examination with the scanning electron microscope, we identified those “hairs” as trichomes which normally grow on the epidermis but not in the eye. Bon overexpression alone can induce trichome formation in the eye which is enhanced by simultaneous Yki overexpression, but we were not sure if this enhancement was due to the overproliferation caused by Yki. So, it was important to determine whether yki loss of function could rescue the trichome phenotype. When I saw that knockdown of yki or sd almost completely wiped out the trichomes, I was so excited and couldn’t wait to email Alexey the images. Alexey replied: “I think we are onto something with the trichomes.” Turned out that he was right, the trichomes ultimately helped us figure out the deeper mechanisms of cell fate determination in the eye.
Another exciting time was when I completed the RNA-seq data analysis and found the downregulation of Notch targets and upregulation of epidermal genes by Bon and Yki. I was very excited as the gene expression is consistent with the biological phenotypes, and these genes could be the target genes we’ve been searching for. Then, when I saw the beautiful eye-to-antenna transformation resulting from overexpression of dominant-negative Notch in the paper [2] that resembled our double antennae phenotype with Yki overexpression or wts knockdown, I thought this is it! And further genetic tests indeed showed that the functions of the Yki-Bon complex are mediated by these targets.
There are also other exciting moments with experiments, such as seeing the epidermal markers and trichomes in wts mutant clone, the gain of eye fate in bon mutant clones, etc. Sometimes the eureka moments came when having discussions with Alexey and Ken, and we suddenly thought of a perfect experiment that could answer the question or solve the problem. And of course, conversations with other people at conferences always brought great inspirations.
4. And what about the flipside: any moments of frustration or despair?
The first couple of years were frustrating. We were trying to find a connection between Bon and the Hippo pathway in the known functions and although we knew Bon and Yki interact, we didn’t know the function of the interaction. In the end, we found exciting new functions that they carry out together. Also, before doing the RNA-seq it was frustrating that we couldn’t identify potential targets from literature reading and data mining. This was resolved after the RNA-seq when we found unexpected non-canonical targets.
5. You used a very broad range of techniques in your paper; did you have any favourites or ones that were more challenging?
I was very lucky to have access to all these resources and have lots of people around me with different expertise who have also been so kind and generous to teach and help me along the way. I love all these approaches, from high-throughput methods and bioinformatics to traditional genetics, as each has its own advantages in answering questions. I found the RNA-seq more challenging at the time since we didn’t have such expertise in our lab before, but it was also an opportunity and I have learned a lot during the process and found myself enjoying deep sequencing data analysis.
6. Where will this story take the lab?
The Veraksa lab is interested in clarifying the exact composition and molecular mechanisms of the multi-protein complexes in which the Yki-Bon module functions. We are also investigating additional functions of the Yki-Bon complex, as well as the functions of Bon and its interactors in various developmental and cellular processes.
7. What is next for you, are you starting a new position?
I have just started my postdoc position with Dr. Oliver Rando at UMass Chan Medical School to study germline and inheritance. I am very excited to explore new areas and expand my skill set.
References:
- Zhao, H., K.H. Moberg, and A. Veraksa, Hippo pathway and Bonus control developmental cell fate decisions in the Drosophila eye. Dev Cell, 2023. 58(5): p. 416-434 e12.
- Kumar, J.P. and K. Moses, EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell, 2001. 104(5): p. 687-97.


 (1 votes)
(1 votes)