BSDB Gurdon/The Company of Biologists 2019 Summer Studentship Report – Lauren Mulcahy
Posted by BSDB, on 18 January 2020
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding. You can read accounts from previous years here. If you’re interested in applying or hosting a student in 2020, applications need to be in by the end of March.
Our fifth report from the class of 2019 comes from Lauren Mulcahy (University of Sussex) who studied fly miRNAs in Sarah Newbury’s lab at the Brighton and Sussex Medical School.
Exploring the genetic control of microRNAs in Drosophila melanogaster
This summer, thanks to the BSDB Gurdon Studentship, I was able to work closely with a PhD student in the lab of Sarah Newbury at the Brighton and Sussex Medical School. Under their supervision I was able to get an applied, practical approach to lab work, much different from that experienced during my undergraduate course.
My project involved looking at the genetic control of microRNAs in Drosophila melanogaster, otherwise known as the common fruit fly. MicroRNAs are small-noncoding RNAs that participate in RNA silencing and regulation of gene expression. miRNAs are able to base-pair with their complementary target mRNA, and through this they are able to silence them, either by their subsequent degradation or prevention of their translation.
Previous studies have shown that exoribonucleases, known as Pacman (XRN1) and Dis3L2 degrade different microRNAs through the mRNA decay pathway, shown in Figure 1.
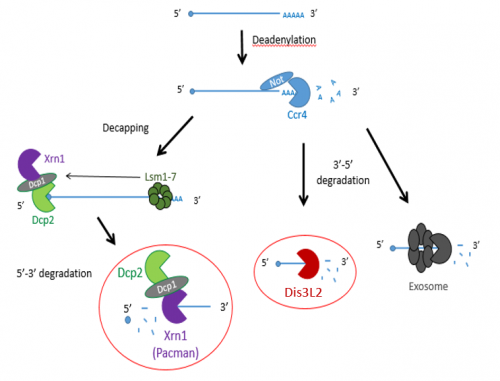
The project made use of wing imaginal discs, which are highly proliferative organs found in Drosophila that will eventually develop into the adult fly’s wings. Prior work in the lab had found that there were significant differences between the size of these discs in both pacman and dis3L2 mutants, with dis3L2 mutants having much larger discs (Towler et al. 2016), and pacman mutants having smaller ones (Waldron et al. 2015). Both exoribonucleases are conserved to humans and their defects have shown to be significant in human disease as well. For example, mutations of DIS3L2 in humans have been linked to Perlman syndrome, an overgrowth syndrome which presents as organomegaly and is associated to a high risk of developing Wilm’s tumours (Astuti et al. 2012). Together this suggested that there could be an involvement of these exoribonucleases in the control and regulation of cell proliferation and apoptosis within these discs, potentially through their degradation activity. Therefore, the aim of my project was to study the difference between levels of specific miRNAs (which were extracted from these WIDs) in both pacman and dis3L2 mutants, compared to their respective control wildtype.
My results, shown in Figure 2, were obtained through the use of qPCR (quantitative or real-time PCR). Also shown on Figure 2 are two other datasets from previous high-throughput RNA sequencing experiments, known as Cuffdiff and sRNAtoolbox.
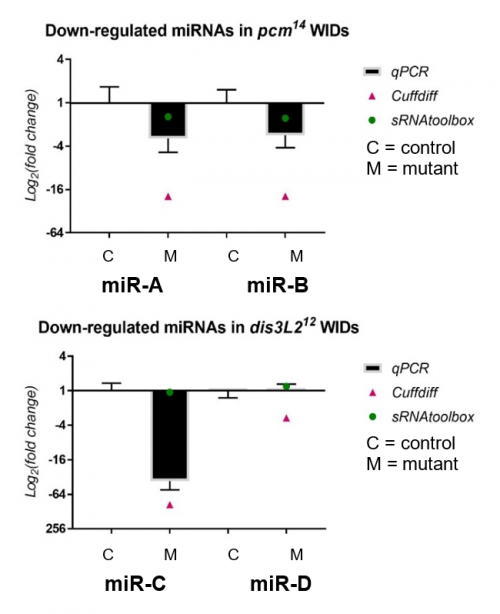
Looking at my data, the pacman mutants showed that there was a negative fold change in the levels of two miRNAs, miR-A and miR-B when normalized to their wildtype’s levels. This indicated that the two miRNAs are downreguated in the pacman mutants. This confirmed the results obtained from the RNA sequencing, as they both showed that the miRNAs were downregulated as well.
Results from the dis3L2 mutants were less definite. For one of the miRNAs, miR-C, there was a negative fold change when normalized to the wildtype’s level. Again, this confirmed results obtained from the RNA-seq, but my data, and data from Cuffdiff, suggested a much stronger downregulation than that from sRNAtoolbox. For the other miRNA, miR-D, qPCR showed that there was no significant fold change in the mutants. This agrees with results from one of the RNA-seq, but not the other. Data from Cuffdiff showed a slight negative fold change of miR-D, suggesting it is downregulated in the mutants. However, my data, and that from sRNAtoolbox, suggested there is no change between the wildtypes and mutants.
Further areas of my project involved looking at the difference in volume of the abdomen and ovaries in flies with a defect in the gene of a TUTase known as Tailor, to see whether it was involved in controlling their size. TUTases, or Terminal Uridylyl Transferases, are enzymes that uridylate miRNAs, potentially serving as a signal for exoribonuclease-mediated degradation such as Dis3L2. This involved me freezing flies in liquid nitrogen, then photographing and dissecting out their ovaries. The limited number of ovaries measured suggested that there were no significant differences in the sizes between the wildtype and mutants.
In addition to this, I also looked at the differences in sizes of wings in Tailor mutant and double mutants of Tailor and Dis3L2 compared to wild types. This included dissecting wings and mounting them on a slide then taking measurements using computer software and a microscope. We hypothesised that the wings from the Tailor mutants would be the same size as the wildtype but the Tailor/Dis3L2 double mutants would have larger wings than the wildtype. My results here were too limited to draw a reliable conclusion but indicated there was no difference in wing sizes between the mutants and wildtype.
Overall, my time in the lab has proved highly valuable and interesting. Learning about the techniques in lectures and actually performing them yourself in a lab are completely different experiences and I am really grateful. I feel it was really important for me to experience the fly work alongside the molecular work as this has helped me to familiarize myself with numerous practises and therefore determine what is most interesting and suitable for me in the future. But not only did I learn how to perform these techniques, I was also introduced into the world of Drosophila, ranging from learning about their genome, recognising their phenotypes, and performing my own genetic crosses to learning their general upkeep and how to do simple things such as egglays or anesthetizing the flies. This experience for me has really increased my interest in developmental biology. Being able to produce my own data and learning to work independently in a lab have also strengthened my desire to do a PhD and I’m very appreciative that I got to experience something not many undergraduate students will. On top of all this, being inside a lab with such a friendly and welcoming group of people has made it all that more enjoyable.
A huge thank you to everyone in the lab for their support and assistance, and to the BSDB for making this opportunity possible through the Gurdon Studentship.
Sources
- Towler BP, Jones CI, Harper KL, Waldron JA, Newbury SF. 2016. A novel role for the 3′-5′ exoribonuclease Dis3L2 in controlling cell proliferation and tissue growth. RNA biology 13: 1286-1299.
- Waldron JA, Jones CI, Towler BP, Pashler AL, Grima DP, Hebbes S, Crossman SH, Zabolotskaya MV, Newbury SF. 2015. Xrn1/Pacman affects apoptosis and regulates expression of hid and reaper. Biology open 4: 649-660.
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ et al. 2012. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nature genetics 44: 277-284.



 (4 votes)
(4 votes)