EmbryoMaker: a general modeling framework to simulate developing systems and perform experiments in silico.
Posted by Miquel Marin-Riera, on 4 October 2015
One of the main challenges of Developmental Biology is to understand the complex developmental mechanisms giving rise to different organs or whole organisms. In most cases, these involve the interplay between cell-cell signalling and cell and tissue movements driven by one or several cell behaviours (such as cell proliferation, cell migration, etc.). Cell signalling will affect how surrounding tissues grow or change in shape, which in turn will change the spatial context in which signalling is taking place. Such complexity in the developmental dynamics can account for the formation of quite complex organs, such as mammalian teeth1 or vertebrate limbs2, but understanding how perturbations on those developmental processes will affect the resulting phenotype is not trivial.
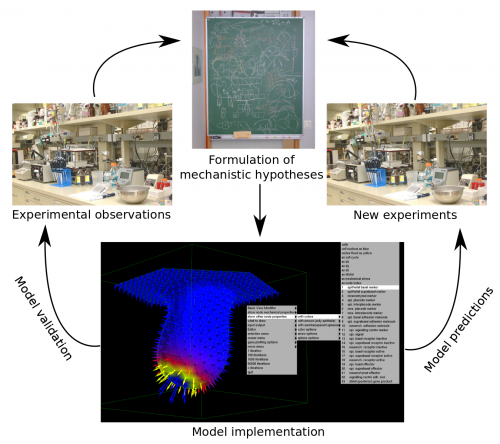
Multi-scale computational models can help in better understanding the dynamics of developmental processes both qualitatively and quantitatively. They should be built upon explicit mechanistic hypotheses about how development takes place. Computational models can then provide explicit quantitative predictions on how the morphology of the organ or embryo or the expression pattern of certain gene products change during development (Figure 1).
In the Salazar-Ciudad lab we work on the design of computational models of development in order to study how the complexity of developmental dynamics may give rise to complex structures and how the presence of different types of developmental mechanisms in different lineages may affect their evolution. For some time we have been using a tooth development model1 in order to approach those questions. However, if one wants to tackle those questions from a more general point of view, organ-specific models of development are not enough. For that purpose we need a modelling framework that implements a general developmental toolkit, that is: 1) the ensemble of cell behaviours known to happen in animal development (cell growth, division, death, cell migration, adhesion, epithelial-mesenchymal transition, cell signalling and extracellular matrix modification), 2) the basic mechanical properties of epithelial and mesenchymal cells and extracellular matrix and 3) the freedom to design gene regulatory networks (GRN) that dynamically control the mechanical properties of cells and their behaviours. Such a modelling tool should be able to simulate the development of, say a tooth or a limb, given that we correctly choose the gene networks and initial conditions in each case. Most interestingly, this would allow to study in silico how to transform one organ into the other by replacing, for example, the tooth forming GRN by the limb forming GRN and vice versa. In a similar way evolutionary transitions between different organs or structures could be inferred by rewiring the GRN step-wise.
Although there already exist modelling frameworks of development3,4,5 none explicitly implements the differential mechanical properties of both epithelial and mesenchymal cells or the whole range of cell behaviours we enumerated above. Thus, we decided to develop the most general developmental modelling framework up to date with all the elements described above.
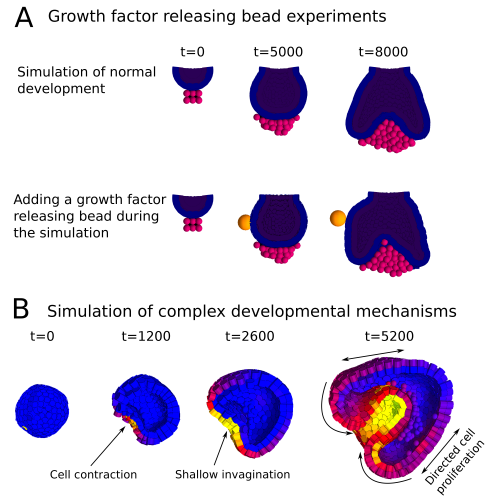
In a paper recently published in Bioinformatics we present this new modelling framework implemented in the open source software EmbryoMaker, freely available for download at our lab’s website. The software provides a graphical interface to visualize the progress of a simulation in real time. In addition, it comes with a user-friendly editing tools in order to design the spatial initial conditions of any developmental system made of either epithelia, mesenchyme and/or extracellular matrix (the size and shape of the tissues at time 0 and their mechanical properties and gene expression profiles) and the GRN (that is defining the number of genes that will participate, their regulatory interactions and their regulation of different cell behaviours). The editing tools can be further used during the simulation of development in order to manipulate the system in real time. For instance, groups of cells can be removed or replaced in the fashion of a graft experiment, or growth factor releasing beads can be placed in any point in the developing system (Figure 2A). Thus, EmbryoMaker may work as an in silico wet lab that allows to predict the outcome of possible experiments on the system of study before carrying them out in vivo or in vitro (Figure 1).
We also show how the modelling framework is able generate complex morphologies from rather simple initial conditions by combining different cell behaviours in a dynamic way (Figure 2B). In this case, the joint action of localized cell contraction, cell polarization and polarized cell division drive the invagination of a spheric epithelium by epiboly.
Overall we expect this new modelling framework to contribute positively to the advance of the Developmental Biology and Evolution fields by providing powerful predictive tools to aid experimental design but also as a means to systematically study the capacity of Development to generate complex and disparate structures and how those might evolve.
Main reference:
Marin-Riera M, Brun-Usan M, Zimm R, Välikangas T, Salazar-Ciudad I (2015) Computational modeling of development by epithelia, mesenchyme and their interactions: a unified model. Bioinformatics
Other references:
1- Salazar-Ciudad I and Jernvall J (2010) A computational model of teeth and the developmental origins of morphological variation. Nature 464, 583-586.
2- Hentschel HGE et al. (2004) Dynamical mechanisms for skeletal pattern formation in the vertebrate limb. Proc. Royal Soc. B. 271, 1713-1722.
3- Pitt-Francis J et al. (2009) Chaste: a test-driven approach to software development for biological modeling. Comput Phys Commun. 180: 2452-2471
4- Izaguirre JA et al. (2004) CompuCell, a multi-model framework for simulation of morphogenesis. Bioinformatics 20, 1129-1137
5- Delile J et al. (2013) Computational modeling and simulation of animal early embryogenesis with the MecaGen platform. In: Kriete A, Eils R editors. Computational Systems Biology, 2nd ed. Academic Press. Elsevier pp. 359-405


 (5 votes)
(5 votes)