Evolution of development and an uncommon model organism
Posted by Erin M Campbell, on 8 December 2010
We can all articulate the importance of using model organisms to understand biology, but many of us fall short in our understanding of some of the more uncommon model organisms. Thankfully, there are amazing biologists out there that save the day! These researchers use some of the more atypical model organisms to understand how different organisms develop and how developmental processes have evolved. Today’s image features the crustacean Parhyale hawaiensis.
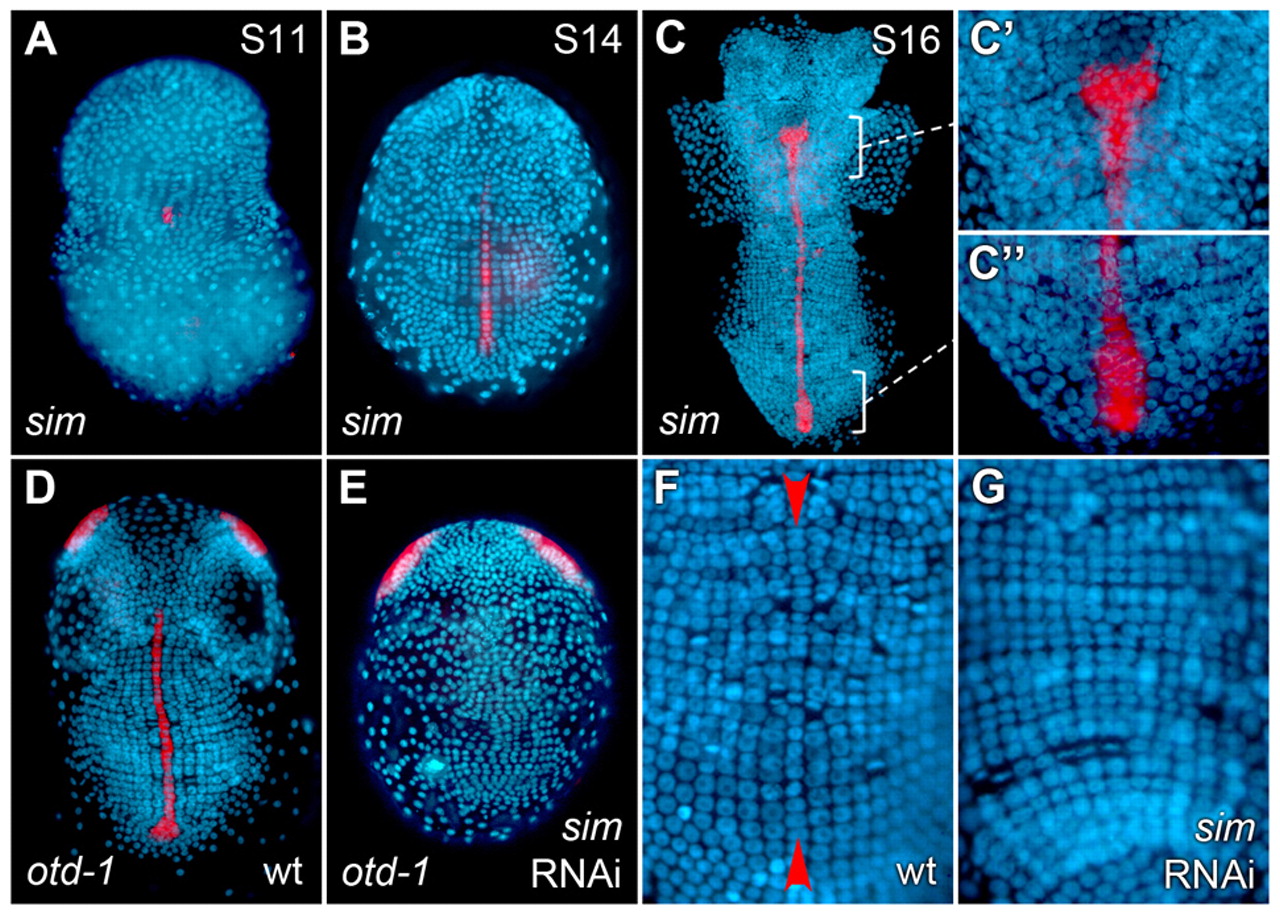
The establishment of the dorsoventral (DV) axis in many organisms is fundamental to the proper organization of organs and tissues. In arthropods, the organization of tissues around the DV axis is well conserved, yet how the axis is established is not. For example, ventral midline cells play a restricted role in DV patterning in Drosophila, yet they play a prominent role in establishing the DV axis in the crustacean Parhyale hawaiensis, according to a recent paper by Vargas-Vila and colleagues published earlier this year in Development. In addition, the Parhyale ortholog of the transcription factor gene single-minded (Ph-sim) is expressed in midline cells and is required for differentiation of midline cells. These results suggest the importance of ventral midline cells in DV patterning in the last ancestor common to both crustaceans and insects.
Images above show Parhyale embryos at different stages, with nuclei in blue. Throughout early development, Ph-sim is expressed in ventral midline cells, as seen in the false-color overlay (red) of expression patterns (A-C). In Ph-sim (RNAi) embryos, midline staining of the midline marker Ph-otd-1 is absent (compare D and E), and the nice clear line of ventral midline cells (red arrows in F) is no longer visible (compare F and G).
![]() Vargas-Vila, M., Hannibal, R., Parchem, R., Liu, P., & Patel, N. (2010). A prominent requirement for single-minded and the ventral midline in patterning the dorsoventral axis of the crustacean Parhyale hawaiensis Development, 137 (20), 3469-3476 DOI: 10.1242/dev.055160
Vargas-Vila, M., Hannibal, R., Parchem, R., Liu, P., & Patel, N. (2010). A prominent requirement for single-minded and the ventral midline in patterning the dorsoventral axis of the crustacean Parhyale hawaiensis Development, 137 (20), 3469-3476 DOI: 10.1242/dev.055160


 (8 votes)
(8 votes)