In Development this week (Vol. 140, Issue 13)
Posted by Seema Grewal, on 11 June 2013
Here are the highlights from the current issue of Development:
Centrosomes and cell fate: a Notch ahead
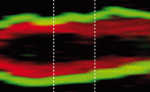 Asymmetric cell divisions (ACDs) play a crucial role in controlling cell fate and generating cell diversity during development. The centrosome is known to be involved in ACD, and recent studies have shown that centrosomes exhibit dynamic and asymmetric movements that regulate orientation of the mitotic spindle. Here, Yohanns Bellaiche and co-workers identify a novel type of centrosome movement during cytokinesis (p. 2657). The authors demonstrate that centrosome movements in Drosophila sensory organ precursors are regulated by the cell fate determinant Numb; the asymmetric localisation of Numb regulates asymmetric centrosome movements. Moreover, they report, Numb acts via the microtubule-binding protein CRMP rather than via its classical effectors. Finally, the researchers show that CRMP in turn participates in the regulation of endosome dynamics and thus likely the recycling of the Notch receptor Delta. They thereby establish a functional link between centrosome dynamics, Notch signalling and cell fate. These findings suggest a model in which asymmetric centrosome movements participate in differential Notch activation to regulate cell fate.
Asymmetric cell divisions (ACDs) play a crucial role in controlling cell fate and generating cell diversity during development. The centrosome is known to be involved in ACD, and recent studies have shown that centrosomes exhibit dynamic and asymmetric movements that regulate orientation of the mitotic spindle. Here, Yohanns Bellaiche and co-workers identify a novel type of centrosome movement during cytokinesis (p. 2657). The authors demonstrate that centrosome movements in Drosophila sensory organ precursors are regulated by the cell fate determinant Numb; the asymmetric localisation of Numb regulates asymmetric centrosome movements. Moreover, they report, Numb acts via the microtubule-binding protein CRMP rather than via its classical effectors. Finally, the researchers show that CRMP in turn participates in the regulation of endosome dynamics and thus likely the recycling of the Notch receptor Delta. They thereby establish a functional link between centrosome dynamics, Notch signalling and cell fate. These findings suggest a model in which asymmetric centrosome movements participate in differential Notch activation to regulate cell fate.
Fishing out maternal and zygotic transcriptomes
 Early embryonic development occurs in the absence of transcription; instead, it relies on maternal mRNAs and proteins present within the egg. It is believed that this period of transcriptional quiescence is maintained by factors that eventually become titrated out during early cleavages, thus leading to zygotic genome activation. How exactly this transition occurs, however, is unclear. Here, Jim Smith, Steven Harvey and co-workers use exome sequencing and RNA-seq to distinguish between maternal and zygotic transcriptomes in early zebrafish embryos (p. 2703). Using single nucleotide polymorphisms to identify maternal and paternal transcriptomes, and using the appearance of paternal mRNAs as an indicator of zygotic transcription, the researchers identify the first zygotic genes to be expressed in the embryo. Zygotic transcription, they report, begins after ten cycles. Prior to this, changes in mRNA levels are observed but these are due to post-transcriptional regulation of maternal mRNAs and not due to transcription. Finally, the researchers demonstrate that different modes of regulation are required for zygotic transcription initiation.
Early embryonic development occurs in the absence of transcription; instead, it relies on maternal mRNAs and proteins present within the egg. It is believed that this period of transcriptional quiescence is maintained by factors that eventually become titrated out during early cleavages, thus leading to zygotic genome activation. How exactly this transition occurs, however, is unclear. Here, Jim Smith, Steven Harvey and co-workers use exome sequencing and RNA-seq to distinguish between maternal and zygotic transcriptomes in early zebrafish embryos (p. 2703). Using single nucleotide polymorphisms to identify maternal and paternal transcriptomes, and using the appearance of paternal mRNAs as an indicator of zygotic transcription, the researchers identify the first zygotic genes to be expressed in the embryo. Zygotic transcription, they report, begins after ten cycles. Prior to this, changes in mRNA levels are observed but these are due to post-transcriptional regulation of maternal mRNAs and not due to transcription. Finally, the researchers demonstrate that different modes of regulation are required for zygotic transcription initiation.
Imaging the neurogenic niche
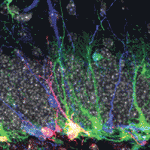 Neural stem/progenitor cells in the mammalian hippocampus generate new neurons throughout life. But how do these integrate into a mature and functional neural circuitry? Here, Sebastian Jessberger and colleagues address this question by using a new imaging approach to analyse neurite growth from newborn granular cells (p. 2823). Using a novel system for culturing sections of mouse hippocampus, combined with retroviral labelling to mark newborn neurons and their progeny, the researchers visualised neurite growth over several days using confocal imaging. Dendritic processes, they report, extended in different directions, with all neurons showing a clear apical extension at ∼4 days. Moreover, the dendrites in such slice cultures follow a linear growth pattern that is characteristic of the growth patterns observed in the intact brain, as assessed by snapshot-based analyses, thus validating their approach. This approach for visualising the adult neurogenic niche opens up the possibility of investigating the dynamic events that occur during adult neurogenesis in both physiological and diseased states.
Neural stem/progenitor cells in the mammalian hippocampus generate new neurons throughout life. But how do these integrate into a mature and functional neural circuitry? Here, Sebastian Jessberger and colleagues address this question by using a new imaging approach to analyse neurite growth from newborn granular cells (p. 2823). Using a novel system for culturing sections of mouse hippocampus, combined with retroviral labelling to mark newborn neurons and their progeny, the researchers visualised neurite growth over several days using confocal imaging. Dendritic processes, they report, extended in different directions, with all neurons showing a clear apical extension at ∼4 days. Moreover, the dendrites in such slice cultures follow a linear growth pattern that is characteristic of the growth patterns observed in the intact brain, as assessed by snapshot-based analyses, thus validating their approach. This approach for visualising the adult neurogenic niche opens up the possibility of investigating the dynamic events that occur during adult neurogenesis in both physiological and diseased states.
A new vein of lumen formation
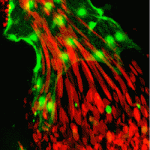 The correct formation of blood vessels is essential for the development of a functional vasculature. Various mechanisms of vascular lumen formation have been described to date but, now, Wiebke Herzog and colleagues examine the development of common cardinal veins (CCVs) in zebrafish and show that these form via a previously undescribed mode of lumen formation (p. 2776). The researchers use in vivo time-lapse studies together with lineage tracing approaches to show that the angioblasts that form CCVs are specified as a population that is distinct from arterial-fated angioblasts. Once specified, these then form CCVs by a novel mechanism, which the authors term ‘lumen ensheathment’: endothelial cells (ECs) delaminate and align along an existing luminal space, extend via migration and eventually enclose the lumen. The delamination and migration events, they report, require cadherin 5, while EC proliferation within developing CCVs requires erythrocyte-derived Vegfc. These findings uncover a new mode of vessel formation, as well as highlighting important crosstalk between the haematopoietic and EC lineages.
The correct formation of blood vessels is essential for the development of a functional vasculature. Various mechanisms of vascular lumen formation have been described to date but, now, Wiebke Herzog and colleagues examine the development of common cardinal veins (CCVs) in zebrafish and show that these form via a previously undescribed mode of lumen formation (p. 2776). The researchers use in vivo time-lapse studies together with lineage tracing approaches to show that the angioblasts that form CCVs are specified as a population that is distinct from arterial-fated angioblasts. Once specified, these then form CCVs by a novel mechanism, which the authors term ‘lumen ensheathment’: endothelial cells (ECs) delaminate and align along an existing luminal space, extend via migration and eventually enclose the lumen. The delamination and migration events, they report, require cadherin 5, while EC proliferation within developing CCVs requires erythrocyte-derived Vegfc. These findings uncover a new mode of vessel formation, as well as highlighting important crosstalk between the haematopoietic and EC lineages.
Specifying hepatopancreas progenitors
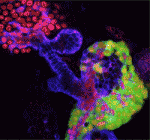 The liver and ventral pancreas are thought to develop from a common pool of multipotent progenitors. Although a number of studies have identified factors required for either pancreas or liver specification, factors that are distinctly required to specify the entire hepatopancreas system have not yet been reported. Now, Joseph Lancman and co-workers uncover a common genetic program, involving hnf1ba and wnt2bb, that specifies progenitors of the liver, ventral pancreas, gall bladder and associated ducts in zebrafish (p. 2669). By characterising a new hnf1ba hypomorphic mutant that phenocopies pancreatic defects found in people with HNF1B monogenic diabetes, the researchers show that hnf1ba regulates pancreas specification and β-cell numbers. Furthermore, they report, the combination of Hnf1ba partial loss with conditional loss of Wnt signalling reveals that these pathways synergize during a narrow developmental window to specify hepatopancreas progenitors; Hnf1ba acts to generate a Wnt permissive domain in the foregut that in turn adopts a hepatopancreatic fate. In summary, these findings highlight a new model for hepatopancreas specification and provide important insights into pancreas and β-cell development.
The liver and ventral pancreas are thought to develop from a common pool of multipotent progenitors. Although a number of studies have identified factors required for either pancreas or liver specification, factors that are distinctly required to specify the entire hepatopancreas system have not yet been reported. Now, Joseph Lancman and co-workers uncover a common genetic program, involving hnf1ba and wnt2bb, that specifies progenitors of the liver, ventral pancreas, gall bladder and associated ducts in zebrafish (p. 2669). By characterising a new hnf1ba hypomorphic mutant that phenocopies pancreatic defects found in people with HNF1B monogenic diabetes, the researchers show that hnf1ba regulates pancreas specification and β-cell numbers. Furthermore, they report, the combination of Hnf1ba partial loss with conditional loss of Wnt signalling reveals that these pathways synergize during a narrow developmental window to specify hepatopancreas progenitors; Hnf1ba acts to generate a Wnt permissive domain in the foregut that in turn adopts a hepatopancreatic fate. In summary, these findings highlight a new model for hepatopancreas specification and provide important insights into pancreas and β-cell development.
A lnc between coding and non-coding RNAs
 Long non-coding RNAs (lncRNAs) have recently emerged as key regulators of gene expression in embryos and in embryonic stem cells (ESCs). Recent large-scale genomics approaches have identified thousands of putative lncRNAs but are these all truly non-coding RNAs? Here, on p. 2828, Alex Schier, Eivind Valen and colleagues set out to answer this question. The researchers use ribosome profiling to identify translated transcripts, combined with a machine-learning approach to classify open reading frames (ORFs) and to validate zebrafish lncRNAs. They find that many proposed lncRNAs are in fact protein-coding contaminants. Moreover, their study reveals that many zebrafish and ESC lncRNAs resemble the 5’ leaders of coding RNAs, suggesting a novel mechanism for lncRNA regulation. Overall, the findings presented here clarify the annotation of lncRNAs, as well as offering a valuable resource that can be used for identifying translated ORFs and hence novel protein-coding genes that function during zebrafish development.
Long non-coding RNAs (lncRNAs) have recently emerged as key regulators of gene expression in embryos and in embryonic stem cells (ESCs). Recent large-scale genomics approaches have identified thousands of putative lncRNAs but are these all truly non-coding RNAs? Here, on p. 2828, Alex Schier, Eivind Valen and colleagues set out to answer this question. The researchers use ribosome profiling to identify translated transcripts, combined with a machine-learning approach to classify open reading frames (ORFs) and to validate zebrafish lncRNAs. They find that many proposed lncRNAs are in fact protein-coding contaminants. Moreover, their study reveals that many zebrafish and ESC lncRNAs resemble the 5’ leaders of coding RNAs, suggesting a novel mechanism for lncRNA regulation. Overall, the findings presented here clarify the annotation of lncRNAs, as well as offering a valuable resource that can be used for identifying translated ORFs and hence novel protein-coding genes that function during zebrafish development.
Plus…
Lineage-dependent circuit assembly in the neocortex
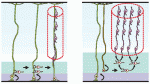 Song-Hai Shi and colleagues review recent findings on the generation, migration and organization of excitatory and inhibitory neurons in the neocortex, and discuss how the lineage history of neurons influences the assembly of functional circuits.
Song-Hai Shi and colleagues review recent findings on the generation, migration and organization of excitatory and inhibitory neurons in the neocortex, and discuss how the lineage history of neurons influences the assembly of functional circuits.
See the Review article on p. 2645
The San Francisco Declaration on Research Assessment
 Read the Editorial by our Editor-in-Chief, Olivier Pourquié on p.2643
Read the Editorial by our Editor-in-Chief, Olivier Pourquié on p.2643
See also the earlier Node post (and some feedback from the community) about this declaration.


 (No Ratings Yet)
(No Ratings Yet)