The evolution of nerves: understanding the roots of neurodegeneration
Posted by Andreas Prokop, on 27 October 2020
Closing Date: 15 March 2021
- Application Deadline: 22 January 2021
- Supervisors: Andreas Prokop, Matthew Ronshaugen, Karl Kadler
- Project details
- How to apply
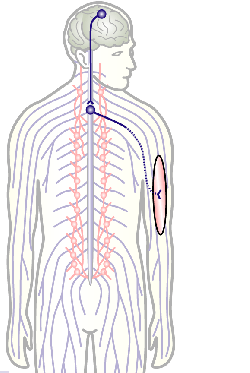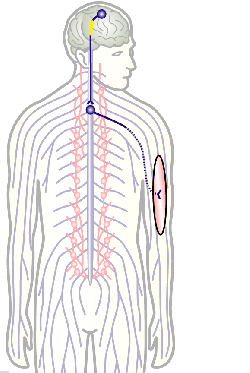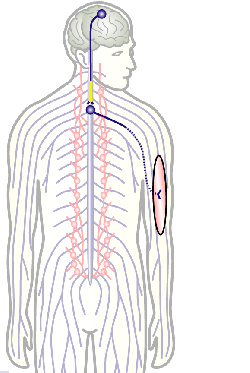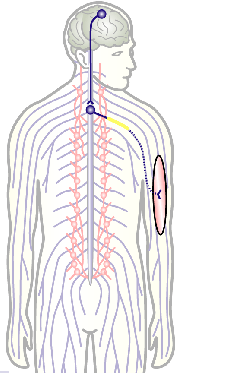
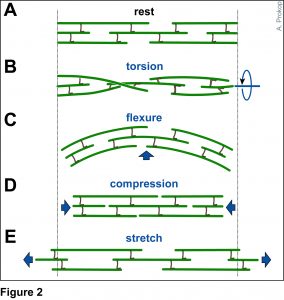
To develop remedial strategies for neurodegeneration in age and disease, we need to improve our understanding of the cell biology of neurons – in particular their axons. Axons are the cable-like, up-to-meter long processes of neurons that wire our nervous system (Fig.1); we lose 40% of axons towards high age and they are key target sites for degenerative processes (Fig.2).
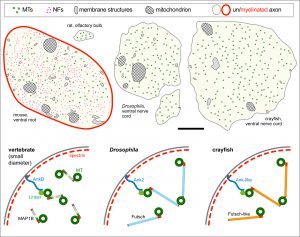
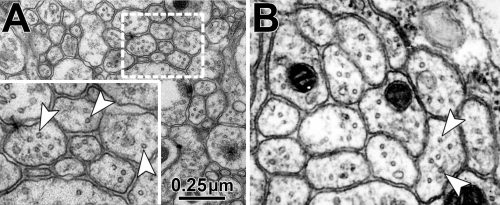
The overarching aim of this project is therefore to gain a better understanding of the architecture of axons and how it evolved from invertebrates to mammals. For this, we focus on the bundles of microtubules (MTs; Fig.3) that extend through the entire axon; they provide the structural backbones of axons and highways for life-sustaining cargo transport and organelle dynamics. Therefore, our studies of the mechanisms that uphold these MT bundles [Refs.1,2] aim to advance our understanding of axon architecture and how they are maintained long-term (Fig.4).
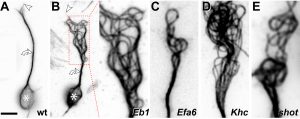
Here we focus on the role of so-called MT cross-linker proteins, expected to hold MTs in parallel bundled arrangements (Figs.3,5). Cross-linkage was one of the first mechanisms proposed to explain axonal MT bundle conformation, and proposed cross-linkers such as Tau or MAP1B have close links to neurodegeneration including Alzheimer’s disease. However, the experimental evidence and molecular understanding of MT cross-linkage is surprisingly sparse [reviewed in Refs. 2,4], leaving an important gap in our understanding of axon architecture and maintenance.
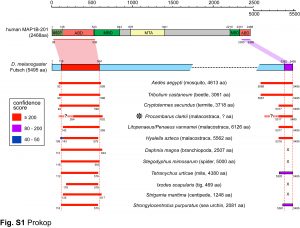
We are particularly interested in MAP1B which plays conserved roles in MT bundle architecture from invertebrates to humans, whilst showing a very particular evolutionary profile (Fig.6): the N- and C-terminal domains have proposed actin-binding properties and are well-conserved throughout the animal kingdom, whereas the middle region is highly variable: it is extremely long (>4,500 aa) in many arthropods correlating with wider spacing of their MTs, whereas other animals including mammals have short forms (1,687 aa in humans) correlating with narrower spacing and the presence of neurofilaments (absent in arthropods). Furthermore, the middle region undergoes rapid change, with sequences not conserved even between closely related species (e.g. Drosophila melanogaster and D. grimshawi flies from the same family). Our detailed studies of MAP1B and its homologues will be doubly beneficial: they will bring important new understanding of mechanisms underpinning evolution, as well as of axon architecture providing information that is also relevant for work on axon degeneration.
During this project, we will capitalise on Matthew Ronshaugen‘s expertise in evolutionary biology to perform phylogenetic analyses of MAP1B and its homologues, aiming to extract concepts and rules that explain MAP1B’s evolutionary behaviour, and develop experimentally testable working hypotheses. Experiments will build on Andreas Prokop‘s expertise on axon structure and MT regulation using Drosophila neurons as a genetically amenable system for fast and efficient experimentation; thus, we will modify the fly MAP1B gene to test our phylogenetics-derived hypotheses. Many experiments will involve electron microscopy for which Karl Kadler is a long-standing expert; EM will reveal structural aberrations, subcellular positions of MAP1B and changes in MT spacing.
Detailed project description
Obj. 1: To understand the evolution of MAP1B homologues: We will perform in-depth evolutionary analysis of the MAP1B protein family by (a) collecting full length MAP1B protein sequences from a broad set of metazoan animal species, (b) building an accurate protein alignment and (c) establishing a stable phylogeny for MAP1B. We will examine how selection has acted on MAP1B domains, for example whether rapid middle region variations are due to neutral drift or possibly positive selection acting to diversify MAP1B crosslinking function and neuronal morphology. Finally, we will use a bioinformatic approach to identify conserved motifs, predict functions based on structure and homology, and examine their gain and loss throughout the MAP1B evolutionary history. Our results will inform the genetic structure function analysis described in Obj. 3 to examine how sequence conservation and divergence within MAP1B proteins correlate with changes in neuronal morphology.
Obj. 2: To establish the mechanisms of Drosophila MAP1B in MT cross-linkage: We will test whether the fly MAP1B is positioned in between MTs and whether it imposes a constant MT-MT spacing in this position. For this, we will perform EM studies using MT contrast enhancement (lanthanum or tannic acid) in combination with MAP1B detectable via a central enzymatic APEX2 tag (generated via CRISPR/Cas9). We know that the conserved N- and C-terminal domains of fly MAP1B are functionally relevant but have no knowledge of the underlying mechanisms [Ref. 3]. We will establish whether they directly bind MTs using biochemical pull-down strategies.
Obj. 3: To functionally assess the meaning of evolutionary changes: Using readouts established during Obj. 2, we will introduce changes to fly MAP1B and assess their impacts on axon structure. We already hold constructs of rat MAP1B and mini constructs of fly MAP1B (containing only N- and C-terminal domains) which will be expressed and tested in MAP1B-deficient background to assess changes, for example in MT spacing. We will build on our phylogenetic analyses and use gene blocks strategies to generate and then analyse interspecies hybrid versions of MAP1B; for example we will assess whether the exchange of the middle region has a structural impact.
Taken together, we will establish how MAP1B proteins contribute to the bundled conformation of axonal MTs across the animal kingdom, and why part of this molecule has undergone such a severe change. This will provide new insights into evolutionary mechanisms and relevant understanding of neurodegenerative processes. This project is therefore highly interdisciplinary and provides training opportunities including phylogenetic in silico analyses, molecular biology, classical genetics, biochemistry and electron microscopy.
References
- Hahn, I., Voelzmann, A., Parkin, J., Fuelle, J. B., Slater, P. G., Lowery, L. A., Sanchez-Soriano, N., Prokop, A. (2020). Tau, XMAP215/Msps and Eb1 jointly regulate microtubule polymerisation and bundle formation in axons. bioRxiv, 2020.08.19.257808 — [LINK]
- Hahn, I., Voelzmann, A., Liew, Y.-T., Costa-Gomes, B., Prokop, A. (2019). The model of local axon homeostasis – explaining the role and regulation of microtubule bundles in axon maintenance and pathology Neural Dev 14, 10.1186/s13064-019-0134-0 — [LINK]
- Hummel, T., Krukkert, K., Roos, J., Davis, G., Klämbt, C. (2000). Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26, 357-370 — [LINK]
- Prokop, A. (2020). Cytoskeletal organization of axons in vertebrates and invertebrates. J Cell Biol 219, e201912081 — [LINK]
- Qu, Y.*, Hahn, I.*, Lees, M., Parkin, J., Voelzmann, A., Dorey, K., Rathbone, A., Friel, C., Allan, V., Okenve Ramos, P., Sánchez-Soriano, N., Prokop, A. (2019). Efa6 regulates axon growth, branching and maintenance by eliminating off-track microtubules at the cortex. eLife 8, e50319 — [LINK]


 (No Ratings Yet)
(No Ratings Yet)