Matching neurons to limbs: an evolutionary perspective on motor system development
Posted by Alana Mendelsohn, on 24 February 2017
Comment on “Divergent Hox Coding and Evasion of Retinoid Signaling Specifies Motor Neurons Innervating Digit Muscles” Neuron 93, 1–14, February 22, 2017.
Alana I. Mendelsohn, Departments of Neuroscience and Biochemistry and Molecular Biophysics, Columbia University
Jeremy S. Dasen, Department of Neuroscience, NYU
Thomas M. Jessell, Departments of Neuroscience and Biochemistry and Molecular Biophysics, Columbia University
The ability of the nervous system to perform complex tasks depends on a diverse set of circuits that underlie specific behaviors. The nervous system generates distinct neural circuits in several ways. One strategy is to make many different types of neurons, varying individual properties such as cellular morphology, settling position, electrical conductance and pharmacological signaling. The other method of forming distinct circuits is to have neurons form selective connections with only a subset of other neurons. Given the scale of the human nervous system, where 100 billion neurons each forms an average of 7000 synaptic connections, the establishment of defined neural circuits is one of the most complex facets of organismal development. The scientific challenge of identifying mechanisms underlying neural specification and circuit formation is in large part what drew me to Tom Jessell’s lab as a graduate student. Tom’s lab works on the mammalian motor system, which carries a number of advantages for studying fundamental principles of nervous system development. Namely, the spinal cord is an excellent model system for studying development because it has well characterized and relatively stereotyped circuit anatomy, with defined sensory inputs and topographically segregated motor outputs.
The first project I worked on in graduate school looked at the formation of monosynaptic reflex circuits between sensory neurons and motor neurons, a process long-thought to be under total genetic control. Although our work showed that the development of these circuits is in fact shaped to some extent by neural activity, it also made clear that one fundamental aspect of circuit formation, the choice of synaptic partner, was made in this context solely on the basis of molecular recognition (Mendelsohn et al., 2015). With this finding in mind, I decided follow up this project by designing a transcriptional screen of different motor pools in mouse, with the intention of identifying molecular surface recognition molecules that might underlie selective circuit formation. Instead, the results of this screen led me down a very different scientific path, towards addressing how motor neurons acquire distinct subtype identities.
In initially following up the results of the screen, I was unable to identify new genes that could plausibly account for differences in sensory-motor connectivity, or even genes that could distinguish motor neurons projecting to different muscles in the leg. But to my surprise, the screen also revealed something completely unexpected: a large number of genes expressed selectively in the motor neurons that project to the intrinsic muscles of the hand and foot. Moreover, I also found that one particularly important gene, Aldh1a2, which encodes the retinoic acid synthesis enzyme Raldh2, was absent in these motor neurons. Given the historical difficulty of defining selective markers of motor pools, this was an exciting and shocking finding.
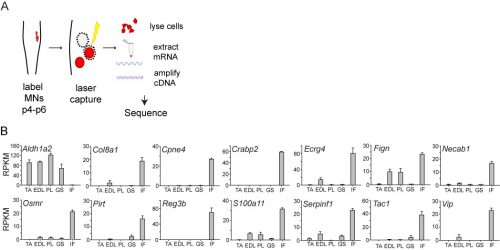
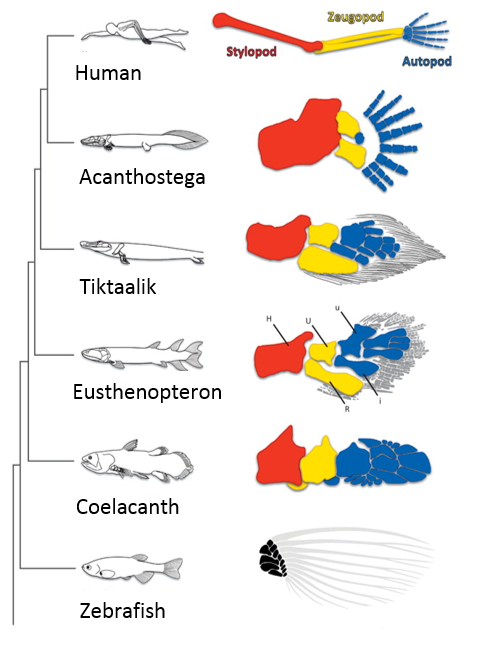
When I first showed these results to Tom, he immediately grasped that they carried profound consequences beyond the scope of developmental neurobiology. The formation of the motor system does not occur in a vacuum, as the development of motor circuits has to correspond with the formation of muscles in the limb that receive motor innervation. In fact, the formation of the limb and motor system use many similar patterning mechanisms, including Shh, FGFs, BMPs, retinoic acid and Hox genes (Cooper et al., 2011; Dasen and Jessell, 2009; Mercader et al., 2000; Roselló-Díez et al., 2011; Zakany and Duboule, 2007). Consequently, Tom realized that the divergent transcriptional profile of digit-innervating motor neurons could have something to do with the natural history of digits themselves. Thinking now in evolutionary terms, Tom quickly sent me off to read Neil Shubin’s best-selling book “Your Inner Fish,” along with a stack of academic papers on, among other things, the embryology of shark and paddlefish fins. I soon learned that evolutionary biologists have long been concerned with the question of how digits formed during the transformation of fish fins into tetropod limbs (Shubin et al., 1997). It appears that the emergence of distal limb elements is a specialized step in limb patterning, requiring evolutionarily conserved molecular determinants (Nakamura et al., 2016; Schneider and Shubin, 2013; Woltering and Duboule, 2010). As a result, we started to wonder whether the specialized evolutionary emergence of digits might explain why we found such divergent patterns of gene expression in digit-innervating motor neurons.
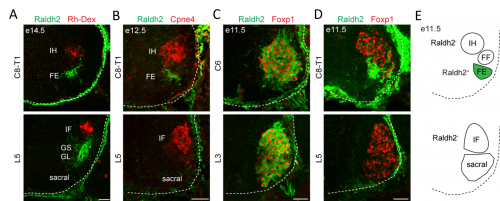
In order to clarify the mechanisms underlying digit-innervating motor neuron specification, we first sought to characterize the patterns of gene expression that emerged from the screen using a combination of in situ hybridization, immunohistochemistry and retrograde anatomical tracing. This work revealed several markers, including Cpne4 and Fign, which selectively labeled digit-innervating motor neurons throughout embryonic development. It also confirmed that digit-innervating motor neurons fail to make retinoic acid from the moment of their specification. In looking at patterns of gene expression under the microscope day after day, I stumbled on another interesting way in which digit-innervating motor neurons are unique. Unlike other limb-innervating motor neurons, digit-innervating motor neurons express the signaling molecule pSMAD in a manner that is dependent on retrograde cues from the limb. Though intrigued by this finding, we weren’t sure how to proceed given that not much is known about the role of the SMAD family in motor neuron development. On the other hand, it was already well known that retinoic acid is involved in the initial generation of motor neurons, as well as their acquisition of broader divisional identities (Sockanathan et al., 2003). As a result, we decided to focus on the role of retinoic acid signaling in digit-motor neuron development.
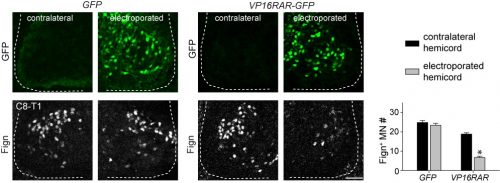
Since digit-innervating motor neurons fail to make retinoids, we wondered whether the absence of retinoic acid was necessary for their development. To address this, we decided to force developing motor neurons to engage in retinoic acid signaling, reasoning that the non-digit innervating motor neurons that already made retinoids would be unaffected. The quickest and easiest way to test this question was to go into chick, where embryologists have long used in ovo electroporation as a technique to manipulate gene expression in developing embryos. We electroporated chicken embryos with a genetic construct in which the retinoic acid receptor RAR was fused to a motif that would render the receptor constitutively active, and found that while other limb-innervating motor neurons were unaffected, the development of digit-innervating motor neurons was selectively disrupted.
The finding that retinoid evasion was important for digit-innervating motor neuron development was satisfying for several reasons. One is that retinoic acid is responsible for the downstream regulation of hundreds of other genes, providing a potential explanation for why digit-innervating motor neurons express so many different genes from other limb-innervating motor neurons. A second reason is that the absence of retinoic acid signaling also helped to explain why intrinsic hand and intrinsic foot innervating motor neurons expressed so many of the same genes, even though each pool is located in a different part of the spinal cord and innervates separate limbs. However, we were still left with the nagging question of why digit-innervating motor neurons were failing to make retinoic acid. To address this, we turned to Hox genes.
Hox genes are involved in both limb and nervous system planning, and have been a longstanding subject of interest in Tom’s lab for their role in specifying motor neurons. Much of this work had been carried out by Jeremy Dasen when he was a post-doc in the lab, in a series of classic studies that I first became acquainted and enamored with as an undergraduate student (Dasen et al., 2003, 2005, 2008). In re-reading these studies, as well as more recent work from Jeremy’s lab at NYU, I started to hone in on the role of two Hox genes, Hoxc8 and Hoxc9. Hoxc8 is expressed at brachial levels of the spinal cord and contributes to limb-innervating motor neuron specification, whereas Hoxc9 is expressed at thoracic levels and is thought to block limb-innervating motor neuron identity. What I found interesting is that even though most limb-innervating motor neurons were unaffected by the loss of Hoxc9, there was a surprising reduction in the number of motor neurons in these mutants projecting to the distal limb (Jung et al., 2010). After initially confirming that digit-innervating motor neurons at brachial levels expressed Hoxc8 as well as low levels of Hoxc9, I started to wonder whether they might both be required for their development. This possibility was especially tantalizing given previous evidence that Hoxc9 is capable of blocking the expression of Raldh2 (Dasen et al., 2003, 2008; Jung et al., 2010), possibly serving as an explanation for why digit-innervating motor neurons fail to make retinoic acid. At this point in the project, it was clear that we would benefit from directly enlisting Jeremy’s help.
We first asked whether Hoxc8 and Hoxc9 were necessary for digit-innervating motor neurons specification, examining the expression of digit motor neuron selective markers in Hoxc8 and Hoxc9 mutants, which Jeremy’s lab provided for us (Catela et al., 2016; Jung et al., 2010). We found that whereas all caudal motor neurons, including those projecting to the hand, were lost in the Hoxc8 mutants, only digit-innervating motor neurons were lost in the Hoxc9 mutants. Moreover, when we ectopically expressed Hoxc8 and Hoxc9 together in chick embryos through electroporation, we could induce the generation of digit-innervating motor neurons, suggesting that this combination of gene expression was also sufficient to specify digit-innervating motor neurons.
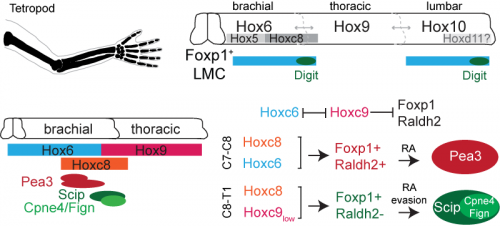
Together, our Hox and retinoid manipulation studies suggested a model for the specification of digit-innervating motor neurons, implicating low levels of Hoxc9 in regulating the absence of Raldh2 in these neurons. While this work provides an important initial step to clarifying how digit-innervating motor neurons acquire their divergent identity, it raises many more questions than it answers. We still don’t understand the role of pSMAD signaling in these motor neurons or how Hox genes operate at lumbar levels of the spinal cord. We also don’t know how retinoid evasion coordinates the expression of downstream genes or the molecular mechanisms by which digit-innervating motor neurons form appropriate connections with muscle in the periphery. But what made this project so exciting and satisfying to work on was that it emerged spontaneously. I had no idea when I started my PhD that I would end up working with chick embryos or reading about the latest advances in paleontology. The freedom to pursue unforeseen and interesting questions and the joy of unexpected discovery has been the most meaningful part of my graduate work, and one that I will hopefully continue to experience throughout my scientific career.
References
Catela, C., Shin, M.M., Lee, D.H., Liu, J.P., and Dasen, J.S. (2016). Hox Proteins Coordinate Motor Neuron Differentiation and Connectivity Programs through Ret/Gfrα Genes. Cell Rep. 14, 1901–1915.
Cooper, K.L., Hu, J.K.-H., ten Berge, D., Fernández-Terán, M.A., Ros, M.Á., and Tabin, C.J. (2011). Initiation of proximal-distal patterning in the vertebrate limb by signals and growth. Science (80-. ). 332, 1083–1086.
Dasen, J.S., and Jessell, T.M. (2009). Hox networks and the origins of motor neuron diversity. (Elsevier Inc.).
Dasen, J.S., Liu, J.-P., and Jessell, T.M. (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933.
Dasen, J.S., Tice, B.C., Brenner-Morton, S., and Jessell, T.M. (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491.
Dasen, J.S., De Camilli, A., Wang, B., Tucker, P.W., and Jessell, T.M. (2008). Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316.
Jung, H., Lacombe, J., Mazzoni, E.O., Liem, K.F., Grinstein, J., Mahony, S., Mukhopadhyay, D., Gifford, D.K., Young, R.A., Anderson, K. V, et al. (2010). Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67, 781–796.
Mendelsohn, A.I., Simon, C.M., Abbott, L.F., Mentis, G.Z., and Jessell, T.M. (2015). Activity Regulates the Incidence of Heteronymous Sensory-Motor Connections. Neuron 87, 111–123.
Mercader, N., Leonardo, E., Piedra, M.E., Martínez-A, C., Ros, M.A., and Torres, M. (2000). Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127, 3961–3970.
Nakamura, T., Gehrke, A.R., Lemberg, J., Szymaszek, J., and Shubin, N.H. (2016). Digits and fin rays share common developmental histories. Nature 1–16.
Roselló-Díez, A., Ros, M.A., and Torres, M. (2011). Diffusible signals, not autonomous mechanisms, determine the main proximodistal limb subdivision. Science 332, 1086–1088.
Schneider, I., and Shubin, N.H. (2013). The origin of the tetrapod limb: from expeditions to enhancers. Trends Genet. 29, 419–426.
Shubin, N., Tabin, C., and Carroll, S. (1997). Fossils, genes and the evolution of animal limbs. Nature 388, 639–648.
Sockanathan, S., Perlmann, T., and Jessell, T.M. (2003). Retinoid receptor signaling in postmitotic motor neurons regulates rostrocaudal positional identity and axonal projection pattern. Neuron 40, 97–111.
Woltering, J.M., and Duboule, D. (2010). The origin of digits: expression patterns versus regulatory mechanisms. Dev. Cell 18, 526–532.
Zakany, J., and Duboule, D. (2007). The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17, 359–366.


 (No Ratings Yet)
(No Ratings Yet)