New phenotype dataset available for embryonic-lethal mouse knockouts
Posted by DMDD, on 1 December 2016

This post originally appeared on Annotations, the DMDD blog.
New image and phenotype data for embryos and placentas from embryonic lethal knockout mouse lines has been made available on the DMDD website today. The knockout data includes the ciliary gene Rpgrip1l as well as Atg16l1, a gene encoding a protein that forms part of a larger complex needed for autophagy. In total we have added HREM image data for 10 new lines, embryo phenotypes for 11 lines and placenta image and phenotype data for 6 lines.
The new data was released at the same time as enhancements to our website, which were described on the DMDD blog. Keep reading to see some highlights from the phenotype data.
DETAILED EMBRYO PHENOTYPES REVEALED
The comprehensive and detailed nature of DMDD embryo phenotyping means that we are able to identify a wide range of abnormalities. In the data released today, a total of 423 phenotypes were scored across 78 embryos. These included gross morphological defects such as exencephaly and edema, but also abormalities on a much smaller scale such as an unusually small dorsal root ganglion, absent hypoglossal nerve and narrowing of the semicircular ear canal.
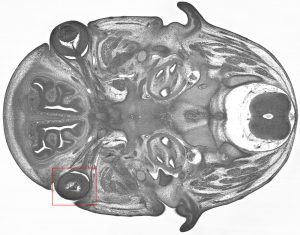
In the image below, a Trim45 embryo at E14.5, was found to have abnormal optic cup morphology and aphakia (a missing lens).
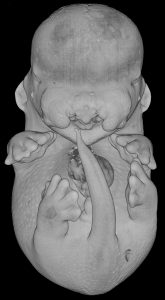
3D modelling of the exterior of an Rpgrip1l knockout embryo at E14.5 revealed a cleft upper lip, as well as polydactyly.
All phenotypes are searchable on the DMDD website, highlighted on relevant images, and the full-resolution image data is available to explore online.
SYSTEMATIC PLACENTAL ANALYSIS
DMDD also carries out systematic phenotyping of the placentas from knockout lines. The image below shows a Cfap53 knockout placenta at E14.5, which was found to have an aberrant fibrotic lesion. The density of fetal blood vessels was also considerably reduced, the overall effect being to reduce the nutrient flow from mother to embryo.

GENE EXPRESSION PROFILES
Work is underway to measure the gene expression profiles for embryos from embryonic lethal knockout lines, a study that complements the morphological phenotype data we are gathering. One of our ultimate goals is to allow data users to explore correlations between gene, morphological phenotype and gene expression profile. The first part of this dataset was released recently – a temporal baseline gene expression profile for wild type embryos.

The expression data is now accessible via a dedicated wild type gene expression profiling page on the DMDD website, which also gives background information about the analysis. Mutant expression data will follow in the new year.
LINKS BETWEEN DMDD GENES AND HUMAN DISEASE
Many of the genes studied by the DMDD programme are known to have links to human disease, including several new lines that have been made available in this release.
Rpgripl1: in humans, mutations in RPGRIPL1 are known to cause Joubert Syndrome (type 7) and Meckel Syndrome (type 5), a rare disorder affecting the cerebellum.
Cfap53: the human ortholog of this gene is known to be associated with visceral heterotaxy-6, in which organs have an abnormal placement and/or orientation.
Arhgef7: in humans the ortholog is associated with Borjeson-Forssman-Lehmann Syndrome.
Arid1b: in humans, mutations in ARID1B are associated with Coffin-Siris Syndrome.
Embryonic lethal lines with no known links to human disease may also be novel candidate genes for undiagnosed genetic disorders. Visit the DMDD website to explore the phenotype data.
A FULL LIST OF NEW DATA
HREM embryo image data has been added for Actn4, Arid1b, Cfap53, Crim1, Cyp11a1, Dmxl2, Fut8, Gas2l2, Mfsd7c, Rala.
Embryo phenotype data has been added for Atg16l1, Capza2, Coro1c, Crim1, Cyfip2, Gas2l2, Gm5544, Rala, Rpgrip1l, Syt1, Trim45.
Placenta image and phenotype data has been added for Arhgef7, Arid1b, Fam21, Fut8, Med23, Timmdc1.
If you have questions about the DMDD programme or our data, please email contact@dmdd.org.uk.


 (No Ratings Yet)
(No Ratings Yet)