The people behind the papers: Nicolas Macaisne & J. Mark Cock
Posted by the Node Interviews, on 1 February 2017
The brown alga Ectocarpus has emerged as a model system for the evolution of muticellularity. Today’s paper, from the current issue of Development, investigates the role and evolutionary history of a gene implicated in Ectocarpus development. We caught up with first author Nicolas Macaisne and supervisor J. Mark Cock of the Station Biologique de Roscoff in Brittany.

Mark, can you tell us your scientific biography and the general focus of your lab?
JMC I am a molecular biologist that worked for many years on various aspects of land plant biology, including nitrogen fixation and self-incompatibility. In 2002 I moved to the Station Biologique de Roscoff, a marine laboratory on the north-west coast of France, with the aim of setting up a laboratory to work on brown algal developmental biology. Working with Akira Peters, we evaluated several brown algae for their potential as model systems and eventually selected the filamentous brown alga Ectocarpus (Peters et al., 2004). The choice of Ectocarpus was based on its amenability to genetic analyses and on earlier work, particularly by Dieter Müller in Constance, that had demonstrated the merits of this species as a laboratory system. The main objective of the laboratory has been to understand the molecular mechanisms that regulate life cycle progression in this brown alga. Ectocarpus has a haploid-diploid life cycle involving an alternation between two multicellular organisms, the sporophyte and the gametophyte. We have been using genetic approaches to identify both key life cycle regulators and regulators of downstream processes during the development of the two generations (Coelho et al., 2011; Peters et al., 2008). In addition, a great deal of effort has been put into generating tools for Ectocarpus, including the sequencing and analysis of its genome in collaboration with the Genoscope in Paris and the VIB in Ghent (Cock et al., 2010). More recently, the laboratory has also become interested in sex determination in the brown algae, a project led by my co-PI Susana Coelho (Ahmed et al., 2014; Lipinska et al., 2015; Luthringer et al., 2015).

And Nicolas, how did you come to work with Mark in Roscoff?
NM I joined Mark’s laboratory in 2011, just after obtaining my Ph.D. under the mentorship of Raphaël Mercier in the National Institute for Agronomic Research of Versailles, France. During my thesis, I studied the genetic regulation of meiotic DNA repair in Arabidopsis, a land plant which is a very well established model for genetics. Thus, my previous research and the model I was employing were quite far removed from Mark’s research on the biology of algae. There are very few labs that study algae, but the unusual life cycle of Ectocarpus, especially the fact that the sporophyte and the gametophyte generations are independent from one another, intrigued me. I realized that this system offered a unique opportunity to study these developmental stages independently. I didn’t appreciate how much I would learn about basic plant biology from this beautiful organism.
I also recognized Mark’s lab as a place where I could challenge myself to learn new techniques and approaches. Working with a model system for which the genome was still not completely assembled and only has a partial genetic map was daunting, but I now feel that I have gained an appreciation for how to set up a new system and look beyond our traditional models for unique insights. This experience was also incredibly valuable for allowing me to learn genome and transcriptome sequencing and analysis. These techniques are absolutely invaluable for the analysis of genes involved in all aspects of developmental biology. Mark’s expertise and the great scientific environment at the Station Biologique de Roscoff are the best assets I could have hoped for in order to achieve my goals.
What makes brown algae interesting models for developmental and evolutionary biologists?
JCM & NM Although many eukaryote groups have evolved simple multicellular forms, we generally consider that only five major groups have evolved complex multicellularity, i.e. possess macroscopic bodyplans with multiple cell types orchestrated by developmental programs (Cock and Collén, 2015). These groups are animals, land plants, brown and red algae and fungi. Complex multicellularity evolved independently in each of these five lineages and comparisons between lineages could therefore provide us with important information about the molecular principals that underlie this key evolutionary transition. Unfortunately, whilst we know a great deal about developmental mechanisms in animals and land plants, the three remaining groups remain almost completely uncharacterised. Of these latter groups, the brown algae are particularly interesting because they include species that rival land plants in terms of their developmentally complexity. We believe that it is important to extend the scope of developmental biology beyond the animal and land plant lineages to effectively address questions about the evolutionary origins of developmental processes associated with complex multicellularity.
“It is important to extend the scope of developmental biology beyond the animal and land plant lineages to effectively address questions about the evolutionary origins of developmental processes associated with complex multicellularity”
The brown algae are also interesting models to address a wide range of more specific developmental questions. Haploid-diploid life cycles, such as that of Ectocarpus, raise the question of how a single genome directs the implementation of two different developmental programs to produce either a sporophyte or a gametophyte at the appropriate stage of the life cycle. Clearly, epigenetic processes must play an important role in this alternation but at present we have no information about how this might be mediated and the epigenetic aspect will be an important avenue for future research. Historically, brown algae have made important contributions to our understanding of early embryogenesis, particularly the establishment of polarity and the asymmetric division of the initial cell (Bouget et al., 1998; Brownlee and Bouget, 1998; Goodner and Quatrano, 1993). In this context, one of the most surprising features of the Ectocarpus life cycle is the alternation between symmetrical and asymmetrical initial cell divisions during the sporophyte and gametophyte generations, respectively. If we look more broadly across the brown algae, these organisms exhibit a bewildering variety of different types of life cycle (Cock et al., 2014) and sexual systems (Luthringer et al., 2014). For example, Fucus species have diploid life cycles and the diecious members of this genus are thought to have diploid phase sex-determination systems (presumably XY or ZW sex chromosome systems) associated with oogamy, whereas the haploid-diploid life cycle of Ectocarpus is associated with a haploid-phase sex-determination system (UV chromosomes) and near-isogamy. Moreover, life cycle and sex-determination systems appear to have varied considerably during the evolution of this group providing an excellent context to link the different types of system with developmental (size, growth habit, reproductive strategy, etc.) and ecological (habitat, biotic interactions, etc.) parameters.
Brown algae also have many interesting features associated with adaptation to the extreme conditions of the coastal environment. These include their unusual cell walls that provide both strength and flexibility, and mechanisms providing resistance to both abiotic (variations in light, heat, desiccation, salinity, etc.) and biotic stresses. Their evolutionary past, which included a secondary endosymbiosis event that gave rise to the stramenopile plastid, provided brown algae with a mosaic of genetic information from various sources, including that of the red algal endosymbiont. Developmental biologists can therefore expect to discover unusual features at the molecular level and the identification of the IMM gene is probably just a foretaste of the novelties to come.
And you appear to have quite an extensive toolkit for Ectocarpus in particular?
JMC & NM Ectocarpus has a long history as a research organism stretching back to the 19th century (Charrier et al., 2008; Coelho et al., 2012). One of our priorities after selecting Ectocarpus as a model organism was to generate a genome sequence and associated genomic and genetic tools including a transcriptomic database, genetic maps and methodologies for investigating gene function. Tool generation is an ongoing process, for example we have recently re-annotated the genome sequence, adding extensive new information and significantly improving the quality of the existing information (Cormier et al., 2016). The Ectocarpus genome is currently one of the highest quality reference sequences available for the stramenopiles. The generation of tools to analyse gene function has been a more complicated process and we still lack one key tool: a robust protocol for genetic transformation. Importantly, the recent development of an RNAi approach, initially for Fucus (Farnham et al., 2013) and subsequently for Ectocarpus (Macaisne et al., 2016), has provided an alternative for reverse genetics and the recent demonstration that forward genetic approaches can be applied in Ectocarpus also represents a powerful tool to analyse gene function. We hope that our demonstration that forward genetic approaches can be applied to brown algal systems will encourage other groups to investigate developmental mechanisms in this group, leading to the emergence of macroalgal developmental biology as a field in its own right.
Can you give us the key results of your paper in a paragraph?
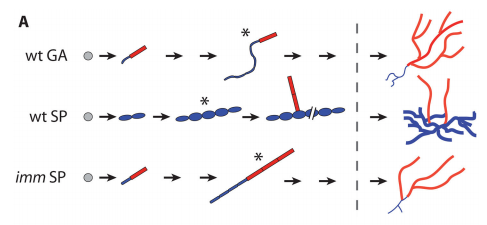
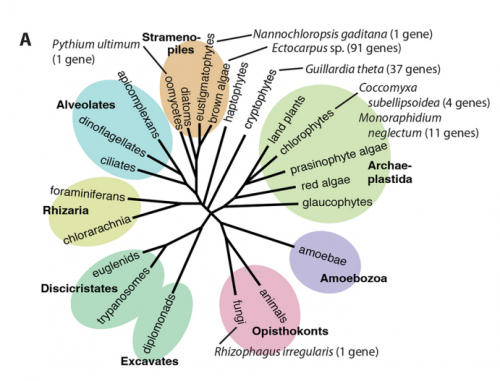
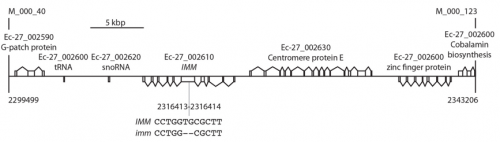
JMC & NM In most brown algae, the initial cell divides to produce two cells with basal and apical identity respectively. This is also the case for the Ectocarpus gametophyte but the sporophyte is unusual because it first establishes an extensive, filamentous basal system before deploying the apical tissues (upright filaments). The first cell division of the sporophyte is therefore developmentally symmetrical, producing the two germ tubes of a symmetrical basal filament (Peters et al., 2008). Sporophytes carrying a mutation in the IMM gene fail to produce the extensive basal system. Instead the initial cell divides asymmetrically to produce a basal rhizoid and an apical upright filament. The mutant sporophyte therefore resembles a gametophyte and the mutation was initially interpreted as causing a partial conversion of the sporophyte into a gametophyte (Peters et al., 2008). In this study, however, analysis of transcriptomic data indicated that the imm sporophyte retains sporophyte identity and we propose an alternative interpretation: that the phenotype of the imm sporophyte represents a more primitive state of the sporophyte, before it evolved the capacity to deploy the extensive basal system. Positional cloning of the IMM locus showed that it encodes a novel protein of unknown function with a repeated cysteine-rich motif that is also found in a gene from the Ectocarpus virus EsV-1. Interestingly, IMM is a member of a large family of 91 genes in Ectocarpus, which we have called EsV-1-7 domain genes after the viral gene that contains this repeated domain. Other stramenopiles either lack EsV-1-7 domain genes or possess only a single EsV-1-7 domain gene suggesting that there has been a massive expansion of this gene family in the brown lineage, perhaps associated with the transition to complex multicellularity (brown algae are the only stramenopile group that exhibit complex multicellularity). A broader search for EsV-1-7 domain genes across the eukaryotic tree of life found a very unusual distribution. In addition to the stramenopile EsV-1-7 domain genes, members of this family were only found in one cryptophyte (Guillardia theta), two chlorophyte green algae (Cocomyxa subellipsoidea and Monorapidium neglectum) and perhaps in one fungal genome (although this latter gene may be a contaminant). It is difficult to reconcile this distribution with vertical inheritance of the gene family. Interestingly, EsV-1-7 domain genes were also found in the genomes of three diverse viruses, suggesting a possible alternative explanation: viral-mediated horizontal gene transfer. This latter hypothesis may be difficult to confirm because horizontal transfer events may have been quite ancient (this appears to have been the case in the stramenopiles for example) but more complete genome sampling for the lineages in question will undoubtedly shed some light on the evolutionary history of this family.
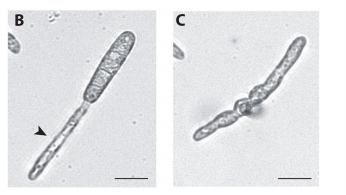
Although IMM seems functionally restricted to the generation of the sporophyte, it is also expressed in the gametophyte: do you think it is doing anything there?
JMC & NM Gametophytes carrying the imm mutation appear to be completely normal indicating that the gene does not have any function during that generation. Of course, we cannot rule out the possibility that there is a subtle or conditional gametophyte phenotype that we have not detected, but the current data indicates that IMM has a sporophyte-specific function. Therefore, in Ectocarpus at least, IMM appears to be specifically associated with the pathway that directs the development of an extensive basal system and delays deployment of the apical system. Nonetheless, the presence of IMM transcripts during the gametophyte generation remains something of a mystery but presumably, if transcription of IMM during the gametophyte generation does not result in any observable phenotype, then there may not have been any selective pressure to prevent IMM transcription during this part of the life cycle.
The IMM gene is conserved in other brown algae, including species that do not exhibit delayed deployment of the apical system. It will be interesting in the future to investigate the role of these IMM orthologues, during both the sporophyte and the gametophyte generations of these other species.
Do you want to hazard a guess at what EsV-1-7 domain proteins are doing?
JMC & NM At present we only have functional information of one member of this family: IMM. The phenotype of the imm mutant suggests this protein has a regulatory function because loss of function leads to changes in the expression levels of a large number of genes. The protein sequence doesn’t tell us very much more about its function except that the repeated cysteine-rich motif in the C-terminal part of the protein (the EsV-1-7 repeat) is reminiscent of a zinc finger motif. Our current guess would therefore be that these proteins contain a novel version of the zinc finger motif that allows them to act as regulatory molecules, perhaps as transcription factors for example. We are currently trying to test this hypothesis by assaying for DNA-binding activity and by attempting to obtain three-dimensional structural information for the protein.
If EsV-1-7 domain proteins have a regulatory role, the next question is what are they regulating? The only information we have at present is their expression patterns in transcriptomic data corresponding to various life cycle stages and conditions. These data, which indicate that the members of the gene family have very diverse expression patterns, suggest that the different members of the family may be involved in diverse processes but additional functional analyses will be necessary to confirm this and to explore the functions of each member of the family.
IMM mutants appear to revert to a more ancestral mode of development, and EsV-1-7 repeat genes may have got into Ectocarpus through horizontal gene transfer. So do you think viral transfer facilitated a new mode of development? It’s a very striking idea!
This would seem to be a reasonable explanation based on the unusual phylogenetic distribution of EsV-1-7 domain genes across the eukaryotes, but it is important to underline that the process would have occurred over a very long time-scale. The presence of EsV-1-7 domain genes in stramenopiles, a cryptophyte, two chlorophytes and perhaps one fungal species, together with the presence of related genes in three viral genomes does strongly suggest horizontal transfer, perhaps mediated by viruses. However, the acquisition of EsV-1-7 domain genes by the stramenopile group appears to be ancient because gene family members were also found in oomycete and eustigmatophyte genomes. Several hundred million years would therefore have separated this ancient horizontal transfer event from the evolution of the IMM gene and the associated novel mode of development in the brown algae.
Nonetheless, we agree that the idea that an important brown algal developmental regulator may have originally been acquired from a viral genome is very interesting. We also believe that the identification of IMM underlines the importance of forward genetic screens and the use of diverse model organisms in developmental biology. None of the classical animal, plant or fungal model organisms possesses EsV-1-7 domain genes, so it would not have been possible to identify this family using these systems.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
Not really a “eureka” moment, but with the hindsight, I realize how lucky I may have been with some experiments I have undertaken. For example, when I was mapping the mutation causing the imm phenotype, after having scanned the genome of 1699 individuals with more than a hundred molecular markers, I was stuck with a relatively large genomic region containing several annotated genes (and possibly more which were not annotated yet) and no easy way to know which of these genes IMM was. So I decided to adapt a whole-genome sequencing approach to sequence specifically this region of the genome of Ectocarpus. Although this approach was successful in identifying the causal mutation, to my knowledge it had never been performed previously, and I was doing that on an alga. So with hindsight, I think I was a bit overly confident, or maybe a bit naive, in thinking that my approach would work. Thankfully it did, and now I am glad I dared to use it.
“With hindsight, I think I was a bit overly confident, or maybe a bit naive, in thinking that my approach would work. Thankfully it did”
And what about the flipside: any moments of frustration or despair?
NM To be honest, I was a bit afraid when I realized the enormity of data that was obtained from the RNA-sequencing, especially considering the fact I had, at that time, no experience in bioinformatics. However, our team and the bioinformaticians of the Station Biologique were extremely supportive; it has really been a team effort.
More generally, my major frustration is that we cannot yet genetically transform Ectocarpus, preventing us from generating another mutant allele of IMM. This would have made the validation of this gene much easier. However, given the successful use of RNAi for this paper, I am confident that transformation tools like CRISPR/Cas9 targeted mutagenesis will be implemented soon for this model organism. Such tools will have a tremendous impact in the field, hastening considerably the understanding of algal biology.
What about your plans for the future following this paper?
NM Working on a relatively young model organism, with a team that is directly involved in the development of genetic and genomic tools to study it, has been very enriching. I have now joined the laboratory of Judith L. Yanowitz at the Magee-Womens Research Institute in Pittsburgh, PA, where I went back to my primary field of expertise, the regulation of DNA repair during meiosis. I now use the nematode Caenorhabditis elegans as a model organism. Now that I have worked on three different organisms, I realized that having a multi-organism approach is highly beneficial for research as we can take advantage of the similarities and the specificities of each organism to better understand a biological process. The next step for me would be to become a principal investigator to continue my research on meiosis using C. elegans as a model and branching out to less well-developed organisms.
“So far we have only scratched the surface and there is still a great deal to be learnt about brown algal developmental biology”
And where next for the lab?
JMC So far we have only scratched the surface and there is still a great deal to be learnt about brown algal developmental biology, including the identification of the major regulators of the diverse processes discussed above. We are still a long way from having any detailed understanding of developmental processes in brown algae. Our laboratory will continue to focus on life cycle regulation and sex determination, using genetic approaches to dissect the underlying pathways but also developing new approaches, for example to investigate the role of epigenetic processes in life cycle control. We are also very interested in the evolutionary perspective and a recently launched project, in collaboration with Genoscope, aimed at obtaining genome sequences for a broad range of brown algal species will provide the genomic data necessary to place brown algal developmental processes in an evolutionary context.


 (2 votes)
(2 votes)