The people behind the papers: Rute Tomaz & Véronique Azuara
Posted by the Node Interviews, on 15 February 2017
Embryonic stem cells express genes necessary for self-renewal, and also ‘prime’ lineage-specific genes which stay silent until differentiation; the molecular players and pathways that govern the timely gene expression are still being delineated. Today’s paper comes from the most recent issue of Development and reveals a role for the histone demethylase Jmjd2c in gene activation in stem cell differentiation. We caught up with first author Rute Tomaz and PI Véronique Azuara of Imperial College London.

Véronique, can you tell us how you first came to be interested in science and how you came to form your lab?
VA Driven by curiosity and the true pleasure of seeing unexpected facets of life really. There is also I think a real satisfaction in “relentlessly” pursuing a question and finding new standpoints to break through it. The question might remain the same the standpoints would vary infinitely. Understanding how a cell identity is defined and sustained is a key question in biology, which I made mine throughout my formative years. First in Paris at the Pasteur Institute studying the ontology of specialized blood cells; then in London at the MRC CSC (now MRC LMS) examining how specific chromatin patterns – fingerprints of a cell identity are established in development initially using haematopoiesis as a model system. Whilst setting up my lab at Imperial College London thanks to a MRC Career Development Award in Stem Cells, our contribution primary focussed on studying the epigenetic regulation of pluripotency and cell fate decisions in the early embryo and derived stem cells.
You’re part of the Stem Cell Regenerative Medicine Network at Imperial, and with the Crick having just opened, London must be a great place for stem cell biology at the moment?
VA Stem cells and developmental biology are indeed key research interests and strengths at the Crick. As an Imperial PI, I will without a doubt nurture collaborative links within this promising science hub in London.
And Rute, how did you come to join Véronique’s lab? I understand you started in Portugal and are now doing a postdoc in Cambridge?
RT That is right. I first stepped into the world of research when I joined Dr Branca Cavaco’s lab (CEDOC, Nova University of Lisbon) for an internship back in Lisbon. There, I studied the molecular aspects of a very rare imprinting disease (Pseudohypoparathyroidism type Ib), and this is what really sparked my interest in epigenetic mechanisms taking place very early on during embryonic development. I then started looking for possible host labs to do a PhD focusing on stem cell epigenetics, and that is when I reached out to Véronique because her scientific interests where a perfect fit for me. In addition, I thought (and still do) that London and the UK are very exciting places to do stem cell research. I was awarded a PhD fellowship from the Portuguese Ministry of Science and Technology, which allowed me to join Véronique’s lab at Imperial where I then explored the role of the histone demethylase Jmjd2c in embryonic stem cells. Stem cells are really fascinating tools, which we here used to mimic embryonic development and take a close look at how gene expression programs are being put in place early on. Over time, this experience has built in me a strong motivation to apply stem cell differentiation methods to the production of relevant cell types for disease modelling. For this reason, I joined Professor Ludovic Vallier’s lab in Cambridge to pursue my post-doctoral training where I am now specializing on hepatocyte differentiation.
Véronique, you’ve worked with embryonic stem cells for more than a decade now. How has the field changed in this time?
VA Stem cell technology is at the heart of regenerative medicine with innovative therapeutic options continually in development. And this was largely made possible by the incredible vitality of basic stem cell research we enjoyed over the past decades; high-quality research investigating fundamental principles and factors that underpin stem cell properties and normal development to provide a reliable foundation for clinical applications.
“Stem cell technology is at the heart of regenerative medicine with innovative therapeutic options continually in development”
There is not much controversy in saying that the area of reprogrammed cells has been a major development in the field. The ability to return somatic cells to an embryonic state is a remarkable breakthrough with the promise of using these cells as tools for translational research. The identification of distinct pluripotency phases, from a naïve to a primed state for differentiation, and characterisation of in vitro growth conditions that would sustain the propagation of these cells is also a key achievement in the field. This brings the prospect of unfolding mechanisms that trigger the exit from naïve pluripotency, enable lineage priming and/or promote specification.
We also greatly benefited from major technology advances including the most recent CRISPR and gene editing technology, as well as the development of single-cell “omic” approaches going hand-to-hand with mounting computational and modelling capacity.
What was known about poised enhancers and lineage priming of embryonic stem cells before your work?
VA & RT Distal regulatory elements such as enhancers have long been recognized to play a significant role in potentiating gene expression. Enhancers are epigenetically delineated by the deposition of H3K4me1/me2 marks with H3K27ac telling apart active (H3K27ac-high) and poised (H3K27ac-low) enhancers, and are commonly bound by pioneer transcription factors. For example, the core pluripotency factor Oct4 was shown to mark both active and poised enhancers in naïve and primed pluripotent stem cells. Enhancer activity and ESC-specific gene expression were also thought to entail long-range DNA interactions with the transcriptional apparatus at promoters, involving the cooperative action of Mediator-Cohesin complexes. Yet, we did not know a great deal about the identity of proteins and mechanisms that stabilize the formation of such assemblies.
Lineage priming occurs at the onset of multi-lineage differentiation of ESCs, and is commonly seen as a “preparation” step for subsequent lineage specification. At the chromatin level, this was typified, for example, by the discovery that many inactive developmental regulators carry bivalent domains being enriched for the repressive H3K27me3 marks along with indicators of permissive chromatin (i.e. early replication timing, methylated H3K4). Bivalent marking was thus proposed to prime lineage-specifying genes for future activation yet prevent their premature expression through Polycomb-mediated repression in ESCs. In trophoblast and extra-embryonic endoderm stem cells, many of the same bivalent promoters would typically acquire additional repressive layers – i.e. Suv39h1-mediated H3K9 methylation and de novo DNA methylation as these genes are not needed for the further development of extra-embryonic tissues.
Can you give us the key results of the paper in a paragraph?
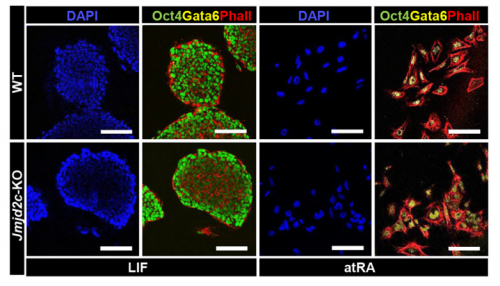
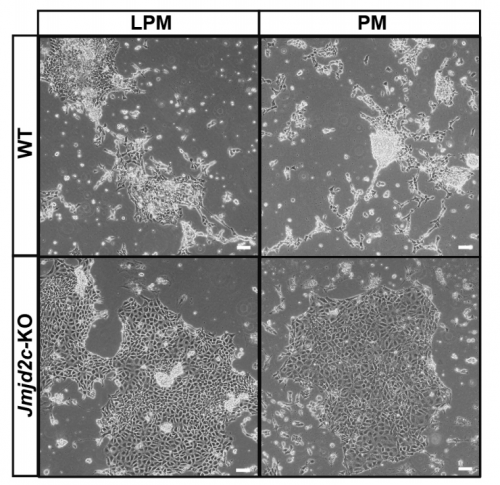
VA & RT We constitutively depleted the H3K9-demethylase Jmjd2c in mouse ESCs and found that this causes an early blockage in multi-lineage differentiation. In contrast, these cells retain the ability to differentiate into extra-embryonic derivatives, where Jmjd2c is not normally expressed. At the chromosomal level, we mapped Jmjd2c binding sites and confirmed that, in our system, Jmjd2c primarily binds to active and bivalent promoters in ESCs. In addition to that, we uncovered that the protein is also timely re-distributed to lineage-specific enhancers at the onset of ESC differentiation. Unexpectedly, we found that Jmjd2c co-occupies active and poised enhancers together with an antagonistic enzyme, the methyltransferase G9a, and that both molecules form a complex with Mediator. Depletion of Jmjd2c abrogates the loading of Mediator-Cohesin and G9a, proposing Jmjd2c as an essential factor for the stable assembly of enhancer multi-protein complexes in ESCs.
Do you know how Jmjd2c itself is being recruited to promoters and enhancers?
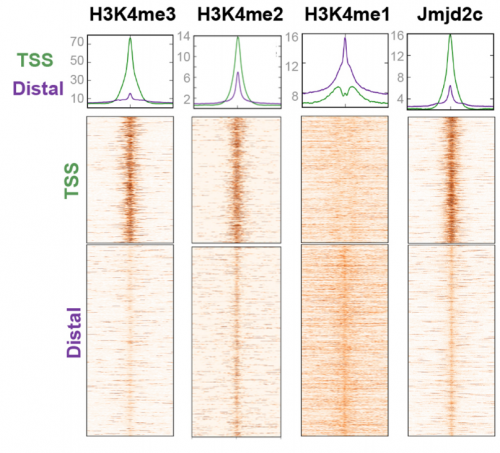
VA & RT We don’t know exactly whether there is a specific transcription factor that recruits Jmjd2c to promoters and enhancers, but what we do know is that two functional Tudor domains and recognition of modified histone tails seem to be important in this recruitment. Published work has shown that Jmjd2c’s Tudor domains can recognise H3K4me3 and H3K4me2 marks, the latter being enriched at both TSS and distal regulatory regions. In our study, we show that deleting these domains abrogates Jmjd2c recruitment at promoters and lineage-specific enhancers. In contrast, this does not impede Jmjd2c binding to active enhancers evoking a different mode of recruitment at these active sites and speculate a possible link with another member of the Jmjd2 family – Jmjd2b. This is mainly for two reasons: Jmjd2b was previously found to bind active enhancers in ESCs; and Jmjd2 family members can form heterodimers, so this could be the case.
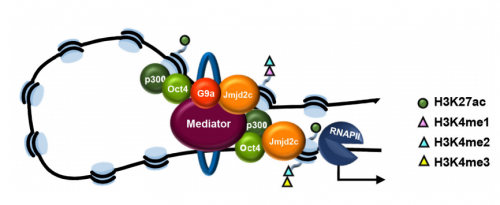
Jmjd2c is co-recruited to enhancers with another histone modifying enzyme, G9a, but you think that they are not acting directly on histone to influence gene expression. So what are they doing?
VA & RT Finding these two molecules together at enhancers but not at promoters was very surprising, most specifically because the levels of H3K9me2 enrichment were not changed at these sites in the absence of Jmjd2c. This made us wonder whether G9a and Jmjd2c play roles beyond their canonical histone modifying activities. We know from published work that, in ESCs, the interplay between G9a and PRC2 safeguards repression of developmental-associated genes. In our work, we find that G9a assembles with Jmjd2c-Mediator-Cohesin at active and poised enhancers. Therefore, we think that G9a might form part of both repressing and activating complexes in ESCs. Interestingly, a dual role for G9a was previously described in hematopoietic cells, where G9a participates in two distinct protein complexes: an “activator” complex with Mediator and a “repressor” complex with Jarid1a, both regulating the differential activation of globin genes in development. The key question is what makes the switch. Published in vitro work has shown that G9a can auto-methylate itself and this mediates the anchor of other repressive factors such as HP1 and Cdyl. A similar link with PRC2 is yet to be explored. On the other hand, non-histone targets have been identified in vitro for the different Jmjd2 family members. Interestingly, these included the auto-methylation site of G9a. One exciting possibility would be that the integration of G9a into distinct complexes relies on its methylation state, and that, in ESCs, this is mediated by Jmjd2c. Being part of an activator complex could indeed require Jmjd2c-dependent demethylation, whereas its methylated form creates a platform for recruitment of repressive complexes.
“Finding G9a and Jmjd2c together at enhancers but not at promoters was very surprising, and made us wonder whether they play roles beyond their canonical histone modifying activities”
When doing the research, was there a particularly exciting result or eureka moment that has stayed with you?
RT There were definitely moments (or results) we recognised as turning points in this work and I will not forget them. We had a very strong phenotypic effect of Jmjd2c-knockout, in terms of ESC differentiation blockage. And throughout this process we were driven by the original hypothesis that Jmjd2c, being an H3K9-demethylase, might be required to “protect” bivalent promoters from acquiring methylation and becoming silenced. I guess a key turning point in this study was the realisation that this might not be the whole story at all. Promoters of lineage-specific genes were not significantly de-repressed in constitutive knockout cells, despite ChIP results showing that Jmjd2c was recruited at these sites. And this is when I started noticing the “little peaks” on Jmjd2c ChIP-seq profiles that seemed always located distally from TSS regions in the “primed” (serum) dataset. By curiosity, we pulled out the published G9a ChIP-seq data and found that this antagonistic enzyme was frequently sitting at these same sites than Jmjd2c. The true excitement came when realising that these sites were actually poised enhancers. I can’t say this was a eureka moment since we still don’t really know what these two are doing together, but it was definitely an exciting one, that opened up some new questions.
And what about the flipside: any moments of frustration or despair?
RT Frustration and despair are part of the process! Luckily we have been supported by a great number of collaborators who shared with us key expertise we were lacking. For example, setting up endogenous co-immunoprecipitation assays was quite demanding and frustrating. Fortunately, Lauriane in Slimane’s lab had the know-how and years of experience and thus was able to perform this set of experiments with ease. I guess the important lesson I learn when in despair is – call for help!
“Frustration and despair are part of the process!”
As an early career stem cell researcher I wonder what you think the stem cell field is going in the next decade?
RT Technology advances exponentially and I think this will have an impact in all sectors of biomedical research. Nowadays we have better methods for genetic engineering, faster and more robust equipment, and the high-throughput sequencing platforms keep evolving significantly. These constant developments will enable us to answer questions in a faster and more profound way, and learn, for example, how signalling and transcriptional networks operate in cellular contexts. This in-depth knowledge we get from stem cells and the developing embryo will have a great impact on our ability to generate conditions that fully mimic in vivo environments on a dish, and recapitulate cell interactions. In particular, I think we will see great advances in 3D tissue culture systems, and the potential of these in disease modelling and drug testing will be limitless.
Where next for the Azuara lab following this work?
VA Surely the lab will continue investigating the epigenetic foundation of pluripotency following-up on Rute’s inspiring study, as well as exploring novel and fundamental links between transcriptional switches, metabolic flux dynamics and cell state transitions in pluripotent stem cells. In fact, we are currently inviting applications from enthusiastic and talented postdoctoral scientists to join us and pursue exciting lines of research at Imperial.
, , , , , , , , , , , , , , ,
Browse the People behind the Papers archive here


 (3 votes)
(3 votes)