The people behind the papers – Dan Dickinson
Posted by the Node Interviews, on 29 September 2017
Cell polarisation is crucial for normal development and controlled by complex molecular interactions in the cytoplasm and at the membrane. Today we feature a paper recently published in Developmental Cell that describes a single-cell biochemistry technique and its insights into polarity protein dynamics the developing worm embryo. We caught up with first author Dan Dickinson, who carried out the work as a postdoc in Bob Goldstein‘s lab in UNC Chapel Hill, and has recently started his own lab in the University of Texas.

Looking back, did you always want to be a scientist?
Well, math and science were always my favorite school subjects, even going back to my elementary school years. I was most interested in astronomy early on – I wanted to be an astronaut in second grade, and most of my early science fair projects involved the solar system or telescopes in some form. I got excited about biology in 7th grade, when we were introduced to Mendelian genetics in a science unit. It was my first introduction to the idea that living things behaved according to a set of knowable rules, and I was hooked almost immediately.
And can you tell us your scientific biography up until now?
My first “real” science experience was the year after I graduated from high school. I was younger than most of my classmates, and so I decided to take a year off before college. I got a job as a tech with a small startup pharmaceutical company that was trying to develop generic versions of several chemotherapeutic drugs. In return for washing dishes half-time, they let me do some small research projects trying to optimize drug yields. It was a fun year and a great window into how corporate research works.
I went to college at Iowa State and joined Gloria Culver’s lab halfway through my freshman year. The lab studied the biochemistry of ribosome assembly in E. coli, and I worked on a couple of different projects related to how one particular protein, called S15, binds and alters the structure of the ribosomal RNA.
During my senior year, I got invited to apply for a Fulbright scholarship, and received an award to go to Switzerland for a year and work in a research lab. I got connected with Martin Pruschy at University Hospital in Zurich, who was studying the signalling pathways that are activated in response to ionizing radiation in cancer cells. My project in that lab was trying to identify novel proteases that were activated by radiation. It was pretty high-risk and the project never really went anywhere, but it was my first time working full-time in an academic lab and I learned a lot about how science really works. It was great preparation for grad school.
I got my Ph.D. at Stanford, where I was jointly advised by James Nelson (a cell biologist) and Bill Weis (a crystallographer and biochemist). I proposed a new project for the two labs, studying the evolution of cell-cell adhesion in the slime mold Dictyostelium. The projected started out because I got curious about evolution, started doing blast searches, and found out that Dicty has homologs of beta-catenin and alpha-catenin, two cell-cell adhesion proteins that we had previous assumed were only present in animals. I wanted to figure out what Dicty was doing with these proteins, and we went in with the hypothesis that it might have something to do with adhesion during the multicellular phase of its life cycle. We were surprised to learn that in fact, during the multicellular phase, Dicty forms a tissue that looks very much like an animal epithelium, and this tissue required the catenins for its structure and polarity. This was a big deal because it suggested that the basic organizational principles behind animal multicellularity might be much more ancient than anyone thought.
I wanted to move into an animal model system as a postdoc, and picked the Goldstein lab after interviewing in several worm and fly labs. I was attracted to the simplicity of C. elegans embryos and the ability to study development at single-cell resolution.
Before this paper, you were among the first to adapt CRISPR/CAS9 in worms. Have you always liked developing new methods?
It’s funny – I can see why someone would ask that after having read my papers (especially from my postdoc). But I don’t think of myself as a “methods guy” at all. I didn’t start working on methods until the second year of my postdoc, and it was only because I got frustrated by the lack of available tools for the experiments I wanted to do. In hindsight, I guess I do enjoy the challenge of working out new techniques, but I’ve always been motivated by the biology.
What do in vivo biochemical methods like the one described in your paper promise to tell us about development?
Well, at the risk of over-generalizing, I’d say that most developmental problems have historically been inaccessible to biochemistry. There are exceptions, of course – Xenopus egg extracts come to mind as a beautiful biochemical system with developmental relevance – but broadly speaking, it’s been hard to get pure populations of cells or tissue from in vivo models in sufficient quantity for biochemistry. The sc-SiMPull approach we developed, combining nanoscale microfluidic lysis and single-molecule biochemistry, represents one solution to that challenge.
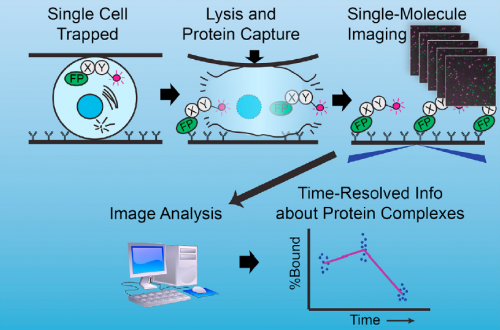
Can you give us the key results of the paper in a paragraph?
There were two really key findings. The first was a demonstration that technically, it is possible to get dynamic information about protein-protein interactions in single C. elegans zygotes by lysing them and performing single-molecule pull-down. Second, by applying this approach, we found that the PAR complex – a critical cell polarity determinant in many systems – dynamically oligomerizes during polarity establishment. We studied the significance of that transient oligomerization using targeted mutants and live imaging, and found that PAR complex oligomerization is essential for PAR proteins to get transported to the anterior of the cell by the cortical flows that establish polarity. We also found that a cell cycle kinase, PLK-1, controlled PAR complex oligomerization by directly phosphorylating one of the complex members (a protein called PAR-3). This was exciting because prior to our work, I don’t think anyone really understood how the timing of polarity establishment was controlled.
How applicable do you see sc-SiMPull being to other systems?
I think it should be straightforward to apply to other systems, and in fact that’s one of the things we’re going to work on in my own lab (which just started this past month, at the University of Texas at Austin). Now that the basic approach is worked out, applying it to other systems should be as simple as adjusting the design of the microfluidic chips to accommodate whatever sample we want to study.
Your paper came out alongside two others – one in the same issue of Developmental Cell and one in Nature Cell Biology. Was there any coordination between the groups in the publication process, and are the results you describe broadly consistent?
I was fortunate to be in touch with the PIs on those other papers, Nate Goehring and Fumio Motegi, before any of us submitted, and we communicated periodically during the review process. I think all three of us had a genuine desire to see everyone get credit for their work – which I believe is the right attitude to have. All three papers reached a similar conclusion about the role of PAR complex oligomerization in polarity establishment, but there’s actually very little overlap in terms of the experiments each group did. That’s an ideal situation, because the papers reinforce each other and increase our confidence that the conclusions are correct.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
The first time I saw single molecules that I’d pulled out of an embryo, that was a big moment when I knew this was going to work. Also, when I first put the oligomerization-blocking mutant embryos on the microscope and saw that they had an interesting polarity defect, that was a big day when it became clear we’d actually discovered something interesting.
And what about the flipside: any moments of frustration or despair?
I actually starting working on sc-SiMPull almost 5 years ago – before CRISPR – and the CRISPR work was kind of a detour along the way. In fact, the whole reason I started doing CRISPR in the first place was because I thought that endogenous gene tagging was a pre-requisite for the kind of quantitative biochemistry I envisioned (we could have counted complexes of overexpressed proteins, but what would we really have learned?). CRISPR turned out to be a much longer detour than I’d expected. In hindsight, it was worth it, of course, but there were some struggles along the way and times when I wondered whether I’d ever get back to the work I actually wanted to be doing.
You’ve just started your own lab in the University of Texas. How are you settling in to Austin, and what do you aim to achieve in the next few years?
Austin is great. It’s a really fun place to live, it’s relatively affordable, and from a scientific standpoint I think the department is phenomenal. I’ve felt very welcome here and am excited to be getting the lab up and running. This is an exciting time because I feel like we finally have the tools for the kind of science I want to do, and I’m looking forward to deconstructing the PAR polarity system at a level of mechanistic understanding that hasn’t been possible previously. We’ll keep working on worms, but I want to expand into some other systems too.
Finally, what do you get up to when you are outside of the lab?
I have two little boys, ages 3 and 5, who are just tremendous fun. I love watching the reactions they get when they tell their friends and teachers that their dad smashes worms for a living. Since we moved here I’ve been spending most of my weekends building various backyard climbing structures for them to mess around on. I also love to cook (and eat), and I compensate for all the cooking and eating by road cycling about 100 miles a week. One of my favorite things about Austin so far is the cycling culture – there are bike lanes everywhere, and they are widely used even in the heat of the summer. It’s been a lot of fun to explore the city and surrounding area that way.
Daniel J. Dickinson, Francoise Schwager, Lionel Pintard, Monica Gotta, Bob Goldstein. 2017. A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization. Developmental Cell. Volume 42, Issue 4, p416–434.
This is #28 in our interview series. Browse the archive here.


 (6 votes)
(6 votes)