Sweetening with a pinch of salt: maximized Cas9 efficiency in zebrafish
Posted by alebur, on 14 June 2016
Alexa Burger, Mosimann lab, Institute of Molecular Life Sciences, University of Zürich, Switzerland.
When I first heard about the “new” genome editing method in early 2013 called CRISPR-Cas9, I thought: “Never ever again will I work with targeted nucleases!” Now it’s mid-2016, we published our approaches to maximize Cas9 effectiveness in zebrafish with Development (Burger et al. 2016), and I happily changed my take to: “Never ever have I seen a system work better than this!” But in all earnest, it was a long (and sometimes bumpy) journey to establish CRISPR-Cas9 in our zebrafish laboratory with its current efficiency and wide-spread applicability. In this blog post, I want to share with you some of our experiences with this method especially for those who are still struggling with getting CRISPR-Cas9 set up in their lab. I want to encourage you to continue working on it – it will work eventually, and we had encountered every possible and impossible problem along the way!
By now, you must have heard of CRISPR-Cas9 for genome editing. In this system, the endonuclease Cas9 is guided to its target location via a short guide RNA (sgRNA or just gRNA, which is the part modified from the original CRISPR repeats) that binds 20 bp of complementary sequence in the genome with its 5’ end and is bound by Cas9 on its 3’ end. By forming this ribonucleoprotein complex (RNP), Cas9 can efficiently cut DNA at your desired locus in the genome of your preferred model organism.
In Spring 2013, I had just moved with my family to Zurich, Switzerland. I had been a postdoc at Massachusetts General Hospital Cancer Center in Boston; as part of the larger Harvard Medical School, I experienced both myself and through collaborators and friends all the ups and downs of using the more complex zincfinger nucleases and later TALENs in zebrafish (so I hope you will relate to my first reaction when hearing about yet another nuclease-based gene editing toy!). Jonas Zaugg, who performed his master thesis in the Mosimann lab, and myself first started looking into CRISPR-Cas9 in autumn 2013 to generate first mutants. As other approaches published at the time (Chang et al., 2013; Hwang et al., 2013; Jao et al., 2013), we injected in vitro-made, capped Cas9 mRNA together with one of the published gRNAs for the zebrafish gene scl24a5, mutants of which are called “golden” due to their resulting yellow-ish pigmentation (Lamason et al., 2005). To be honest, we initially obtained rather lousy results, struggled with the long Cas9 mRNA and the tiny sgRNAs, and I thought to myself “here we go again…”.
Our genome editing game changed completely once we teamed up with Martin Jinek’s laboratory: Martin had done key work on the Cas9 protein itself during his postdoc in Jennifer Doudna’s laboratory at Berkley (Jinek et al., 2012; Jinek et al., 2013), and he had started his lab on-campus around the same time as our PI Christian Mosimann. The two PIs met at a lunch event, and started chatting about their mutual interest in electric string instruments and science. This led to Martin and his graduate student Carolin Anders dropping by our lab with a liquid nitrogen container containing small tubes of (what we call) crystal-grade Cas9 protein that was either GFP- or mCherry-tagged.
Previous accounts of Cas9 protein use in zebrafish were highly encouraging (Gagnon et al., 2014). Using Carolin and Martin’s input on how they treated Cas9-sgRNA complexes in vitro and for structure analysis, we assembled 850-1000 nanograms per microliter of Cas9 with sgRNA into ready-to-use RNP complexes (mixing and incubating at 37°C for 5min, then kept at room temperature) and injected those into zebrafish embryos. A key ingredient turned out to be sufficient KCl salt to keep Cas9 happy with at least 300mM. As documented in our paper, leaving out this crucial step causes the Cas9 RNPs to clump up quickly in the injection needle, so we highly recommend checking for sufficient salt in your final injection mix! The resulting solubilization is applicable to any Cas9 source and should also work with commercial protein stocks (given it is sufficiently concentrated and has an NLS, which is not always the case…).
The subsequent first experiments were, in retrospect, quite funny. We attempted to mutagenize EGFP, which we have abundantly available in our lab’s many transgenics. After the first EGFP targeting injection into homozygous ubi:EGFP embryos, we didn’t see any fluorescence in the embryos the next day. Thinking we messed up the cross, we repeated the experiment and this time we kept some uninjected controls – these were glowing green as usual the next day, but almost none of the Cas9 RNP-injected embryos showed any fluorescence. That’s when it dawned to us that we had mutated the EGFP ORF with such efficiency that there was no functional EGFP protein whatsoever.
Instantly, we could see the difference in all our other CRISPR-Cas9 targeting using our new tricks. And what a difference that was! From about 5 percent (tops) with Cas9 mRNA to up to 100 percent mutagenesis efficiency with the RNPs. We went on to scrutinize every single step in our sgRNA production, injection protocols, genotyping procedures, etc. Along the way, we made all the mistakes one can make: degraded RNA due to dirty pipettes, wrong storage of Cas9 protein stocks, wrong primer sequences ordered (beware of the reverse-complement!), miscalculated injection mixes…the list goes on. All this was topped in the sheer load of sequences we accumulated of targeted loci that we initially needed to analyze by hand. Another key enemy became the frequent single-nucleotide polymorphisms (SNPs) in the zebrafish genome: single SNPs in a 20bp stretch for sgRNA targeting usually abolishes all detectable mutagenesis in our hands, so we frequently sequence targeted loci before injections.
As a lab, we got together and defined some ground rules for our mutagenesis work. We developed the policy to test every new sgRNA on a denaturing MOPS gel. We asked our bioinformatics collaborators in Mark Robinson’s lab next door to code a simple online interface to calculate injection mixes that Lukas Weber coined CrispantCal, which Raul Catena subsequently also turned into a smart phone app. We developed a cloning- and purification-free sequence verification strategy for routine mutagenesis analysis and collected samples for a massive deep-sequencing verification of 40+ loci.
Most importantly, Helen Lindsay from the Robinson lab developed the R-based software tool CrispRVariants for rapid sequence analysis of mutation events (Lindsay et al., in press; preprint at http://biorxiv.org/content/early/2016/03/10/034140). In simple terms, CrispRVariants uses a smart procedure to align sequence results to the intended target locus and performs a base-by-base comparison to the wildtype reference sequence. Paired with sequence counts, CrispRVariants spits out so-called panel plots that visualize the mutagenesis results with allele details. Continuously refined through our day-to-day experiences, these panel plots became a daily discussion point in the lab as we now had a standardized visualization of mutagenesis events – and we hope the online version CrispRVariantsLite will help also you to easily grasp your mutation spectra!
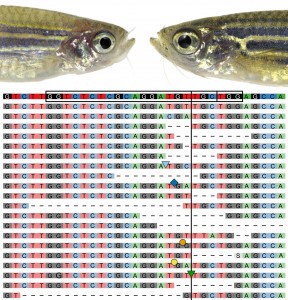
CrispRVariants-based analysis rapidly corroborated that our new protocols achieved exceedingly high mutagenesis efficiencies, reaching even complete biallelic mutagenesis in individual zebrafish embryos. We also started to see previously described loss-of-function phenotypes in injected F0 embryos: we call such Cas9-based somatic mutants “crispants” in analogy to morpholino-injected embryos referred to as morphants. While we do frequently use crispants to figure out if a candidate gene is interesting enough to follow-up as stable mutant in the course of our lab’s work on mesoderm cell fates, we quickly became cautious about using this approach for actual loss-of-function analysis: sequence analysis revealed that most mutated loci have a high propensity to be repaired in a stereotypic set of alleles, including in-frame alleles or even other still functional alleles possibly involving also clonal selection during development. Such effects paired with still variable mutagenesis efficiency make crispant phenotypes for most genes too variable for reproducible, reliable phenotype studies, but nonetheless allow a first glance at the full-blown loss-of-function phenotype. We are harnessing this effect now for limited candidate gene screens for novel cardiovascular regulators and tumor modifiers, and (if space permits!) we alwaysgenerate stable germline mutants for each gene with interesting phenotypes. So beyond tool development, the focus of our endeavors was to be able to probe for new, exciting biology, and we are all enthusiastically applying our new tools to our individual projects.
But keep in mind: realistically, how often do you really need complete mutagenesis? High mutation load in injected animals can perturb their proper development, and you consequently never get adult founders. More important is that, once the Cas9 RNP mutagenesis is optimized, the injections can be titrated to allow for proper development and subsequent screening of germline transmission.
My favorite part of this journey was the collaborative work with protein biochemists, biostatisticians, and our lab’s creative team of biologists. Several spin-off projects have developed and are ongoing in the lab, and we have instructed several outside collaborators in our techniques with hands-on demos during lab visits. Technically, our protocols should be directly applicable to any model organism that allows for injection-based delivery of Cas9 RNPs. In the end however, all comes down to understanding your genetics: once the mutagenesis works, you need to be fluent in the language of classic genetics and quirks such as maternal contribution, phenotype expressivity vs penetrance, hypomorphs, etc. CRISPR-Cas9 is only the beginning. Happy crispring!
Web Links:
CrispantCal web interface: http://imlspenticton.uzh.ch:3838/CrispantCal/
CrispantCal for iPhone: https://itunes.apple.com/nz/app/crispantcal/id1112401634?mt=8
CrispantCal for Android: https://play.google.com/store/apps/details?id=com.raulcatena.crisprcas9
CrispRVariantsLite: http://imlspenticton.uzh.ch:3838/CrispRVariantsLite/
References:
Burger, A., Lindsay, H., Felker, A., Hess, C., Anders, C., Chiavacci, E., Zaugg, J., Weber, L. M., Catena, R., Jinek, M., et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes.
Chang, N., Sun, C., Gao, L., Zhu, D., Xu, X., Zhu, X., Xiong, J.-W. W. and Xi, J. J. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23, 465–472.
Gagnon, J. A., Valen, E., Thyme, S. B., Huang, P., Ahkmetova, L., Pauli, A., Montague, T. G., Zimmerman, S., Richter, C. and Schier, A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9, e98186.
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., Peterson, R. T., Yeh, J. R. and Joung, J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31, 227–229.
Jao, L.-E., Wente, S. R. and Chen, W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U. S. A. 110, 13904–9.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80-. ). 337, 816–821.
Jinek, M., East, A., Cheng, A., Lin, S., Ma, E. and Doudna, J. (2013). RNA-programmed genome editing in human cells. Elife 2, e00471.
Lamason, R. L., Mohideen, M.-A. P. K., Mest, J. R., Wong, A. C., Norton, H. L., Aros, M. C., Jurynec, M. J., Mao, X., Humphreville, V. R., Humbert, J. E., et al. (2005). SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–6.
Lindsay, H., Burger, A., Felker, A., Hess, C., Zaugg, J., Chiavacci, E., Anders, C., Jinek, M., Mosimann, C. and Robinson, M. D. (2015). CrispRVariants: precisely charting the mutation spectrum in genome engineering experiments. Cold Spring Harbor Labs Journals. http://biorxiv.org/content/early/2016/03/10/034140


 (5 votes)
(5 votes)
Nice article. I can’t find the CrispantCal on Android in the Google Play Store, or by using the link above however. Is it specific to an older Android version, but not the latest one?
True! I will inquire about it. Can you use in the meantime our CrispantCal web interface?
Great article!
“A key ingredient turned out to be sufficient KCl salt to keep Cas9 happy with at least 300mM”
Is there a maximum salt concentration that can inhibit Cas9 activity?