The people behind the papers – Andrew Economou and Jeremy Green
Posted by the Node Interviews, on 31 January 2021
This interview, the 89th in our series, was published in Development last year.
Interacting morphogens produce periodic patterns in developing tissues. Such patterning can be modelled as reaction-diffusion (RD) processes (as originally formulated by Alan Turing), and although these models have been developed and refined over the years, they often tend to oversimplify biological complexity by restricting the number of interacting morphogens. A new paper in Development reports how perturbation analysis can guide multi-morphogen modelling of the striped patterning the roof of the mouse mouth. To hear more about the story, we caught up with first author Andrew Economou and his former supervisor Jeremy Green, Professor of Developmental Biology at King’s College, London.

Jeremy, can you give us your scientific biography and the questions your lab is trying to answer?
JG: As an undergraduate I started off studying physics but switched to biochemistry, and I was lucky enough to take the famous Developmental Biology course at Cambridge taught by such luminaries as John Gurdon, Michael Ashburner and Peter Lawrence. That was really what inspired me to follow the subject. For my PhD I got slightly side-tracked into gene expression in yeast, but that was enough to convince me that what I was really interested in was the macro-spatial aspect of biology. I managed to get myself funded to go to a conference where I saw Jim Smith, then a newly-independent investigator, announce his discovery of a mesoderm-inducing factor in Xenopus, and I knew that this was a big deal. Fortunately, Jim took me on as a post-doc and together we did the experiments that showed the inducing factor to be a classic Wolpert French Flag-style morphogen, triggering different cell fates according to dose thresholds.
I then worked with John Gerhart and Ray Keller at UC Berkeley, mostly following up threshold-sharpening with John, but all the while soaking up the morphogenesis culture of the Keller lab. In my own lab (at the Dana-Farber Cancer Institute, part of Harvard Medical School) I then got into cell polarity and Wnt signalling – becoming quite biochemical – but on returning to London and joining King’s College London I re-focused back onto spatial organisation, both with morphogen action and physical morphogenesis.
It has been great harking back to my undergraduate days by approaching this with a physics mind-set, especially working with great collaborators and a mathematically talented postdoc like Andrew.
And Andrew – how did you come to work in Jeremy’s lab and what was the main drive behind your research there?
AE: As a PhD student, I worked in the field of Evolutionary Developmental Biology, looking at the evolution of arthropod segmental identity. Having never had a formal developmental biology training – in my undergraduate I largely specialised in palaeontology – I always felt that there was something missing from the way I thought about embryonic development. After my PhD, I decided that I needed to get a better understanding of the processes that actually shaped the embryo, and it was fortunate that at that time Jeremy was advertising a project for studying the cellular mechanisms underlying the morphogenesis of the mouse face.
When I started with Jeremy, I had no background in any sort of quantitative or computational biology, but it became clear that these approaches were necessary to understanding how the embryo is patterned and shaped. Fortunately, Jeremy was very happy for me to have a go and try to apply computational approaches to my data and see how far we could get. Ultimately, my research became focussed on extracting quantitative information on the different cellular- and molecular-level processes and understanding how they contribute to the distinctive patterning processes seen in the mouse palate epithelium, at the level of both tissue growth and signalling networks.
How has your research been affected by the COVID-19 pandemic?
AE: Like so many other people, I had to put all experiments on hold for the months we were in lockdown. While that obviously held back many parts of my current project (I have moved on from Jeremy’s lab to another postdoc), I have always had some more computational aspect to my work, which I usually keep running in the background. The enforced break from the lab gave me the opportunity to really push on with these parts of my project, so I had plenty of work to keep me occupied! I had also been guilty of allowing a lot of data analysis to build up, so time away from the lab finally gave me a chance to catch up with that. In many ways, this actually helped me understand where I was in my project, and has given me a clearer idea of what my priorities should be since getting back into the lab.
JG: My lab at King’s was shut down completely for a longer time than in some places – partly because we are embedded in a hospital building – so although it has been good for me to catch up on manuscripts, both postdocs and students in the lab are only now (in September) really beginning to pick up momentum again. We were very much in an experimental rather than computational or data analysis phase, so we’re now going to be dependent on supplementary funding from UKRI and King’s to be able to make up for lost time. Fortunately, there are signs that this will be forthcoming, so I’m optimistic.
What have been the main developmental insights – and limitations – of RD modelling before your paper?
AE & JG: Although Turing published his seminal paper almost 70 years ago, we think it’s fair to say that the developmental biology community has only really started to appreciate its importance relatively recently. While we have known for a long time that RD systems can capture biological patterns such as tiger stripes, it has only been in the last 10-20 years that developmental systems have been identified for which RD processes can not only recapitulate the observed patterns, but also for which specific biological molecules have been identified as morphogens.
There has also been a lot of recent theoretical progress, moving beyond the well-established criteria for pattern formation in two-component RD systems, exploring more generally the criteria for the formation of periodic patterns, and looking at how different features of network topology affect the spatial pattern for these larger – more biologically realistic – networks. However, although these larger networks capture more of the complexity of these real biological systems, one thing coming out of these studies was that many networks can produce the same pattern. This is one of the main aspects our paper addresses: how can we use other forms of data, such as inhibiting the different morphogens and following their dynamics during pattern formation, to constrain our understanding of the patterning network?
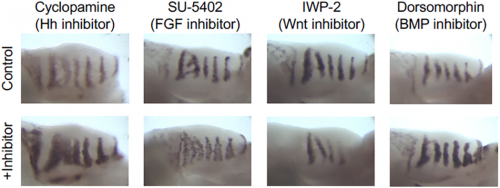
Can you give us the key results of the paper in a paragraph?
AE & JG: RD systems are brilliant for describing the self-organising behaviour of morphogens and morphogen-like systems. RD models, from Turing to the very recent past, have been very focused on the minimal two-component system. (Turing’s genius was to strip problems down to the bare minimum and RD is a perfect example of that: two components is all you need.) But biology is messy and evolution isn’t always parsimonious, so in real life there are often more components than you need. As biologists, we want to explain what’s there and see how all these potentially redundant components fit in. Although two-component RD systems have been extensively studied, it is not clear how such simple systems relate to ‘real’ biological networks. We set out to determine how the four signalling pathways Hedgehog, FGF, Wnt and BMP could interact to produce the pattern seen in the palatal rugae – ridges in the roof of the mouse’s mouth. We first established a relatively simple set of experimental observations: how does the pattern respond to the inhibition of each pathway, and what are their spatial patterns of activity? That showed that FGF was working differently in different tissue layers, so we had five rather than four morphogens to deal with. Then, through numerical simulations and mathematical analysis, we established a general framework determining how the response of one component to the inhibition of another is dependent on their position relative to the feedback loops in a minimal RD network, as well as their relative phase. Combining these constraints with temporal information about the sequence of signalling events during the establishment of a rugal stripe, we were able to identify a set of just 154 topologies out of a total set of 39,755 possible networks involving the four pathways, which were consistent with our experimental observations, constraining the sign of all possible interactions in the network.
Biology is messy and evolution isn’t always parsimonious
Rugae stripes appear to be a fairly simple pattern – why do you think it takes five interacting morphogens to make it, when (I guess) two might do the job just as well?
AE & JG: There are three main reasons we can speculate about. One is that the regulatory networks for this system were in effect borrowed from an existing gene regulatory toolkit that happens to include all of these morphogens. There might not be strong selection to get rid of any, so there they are.
The second is that multiple signals might be needed downstream of the stripe formation as such but the system saves time and maybe other kinds of complexity by building them all into the stripe formation. Ruga morphogenesis involves the formation of an epithelial placode and condensation of the underlying mesenchyme, as well as reciprocal signalling between these tissue layers. We suspect that the feedback generated by some of these processes underlies the incorporation of additional pathways into the network after the ruga stripe is initially formed by a subset of pathways.
A third intriguing possibility is that increasing the number of morphogens may increase the robustness of the system. Recent work from other labs has shown that different network topologies vary in their robustness. Therefore, it is not unreasonable to think that initiating a stripe with a system of three morphogens could, for some topologies, be more robust than one made of just two. Although more morphogens provide more targets for perturbations in levels, we were struck by how few of our in silico perturbations changed the stripe spacing and number, so maybe that’s where the robustness needs to be.
How generalisable do you think your approach will be for other developmental problems? And do you have any advice for researchers who wish to increase the number of components in their RD modelling?
AE & JG: Given that our experimental approaches were based on very standard developmental biological techniques – in situ hybridisation and the application of small molecular inhibitors – there is no reason that this approach cannot be applied to other developmental systems patterned by RD processes. Moreover, we think this approach is quite generalisable, even to systems that aren’t obviously periodic. It is known, for example, that Nodal and Lefty proteins constitute a self-organising Turing/RD morphogen pair for left-right patterning in early vertebrates. (The left-right gradient that’s set up is, in effect, a quarter-wavelength of what would be a periodic system if there were enough tissue to encompass it.) So almost any gradient system that involves self-organising feedback might be analysed in much the same way.
One of the most exciting insights from our work, which could be of interest to other researchers, came from looking into the dynamics of stripe formation. Considering the sequence of events which occurred during the formation of the rugae, alongside the feedback we had identified in the system, helped us realise that we can learn more from such a patterning process than just inferring a static depiction of the interactions shown in the network. Different morphogens in a network have different functions: for example, in our system only a subset of the morphogens is actually involved in initiating a new stripe. As the number of components in a network increase, it is important to consider their function in the network.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
AE: The moment that most sticks with me is when I ran my initial set of simulations to look at the effect of inhibiting each component for our set of minimal networks. I had expected that this analysis would pick out a subset of networks but had never really thought about what this would tell us more generally about our system. On seeing the results – that the response of the system to inhibiting different components correlated with the feedback loops they were in – everything made so much more sense and suggested a framework for thinking about much larger networks than the two- and three-component RD systems we were working with.
And what about the flipside: any moments of frustration or despair?
AE: Having run those first simulations and had the initial insight, we then needed a way to actually demonstrate that these results were applicable not just to the two- and three-component systems for which we had run our simulations, but were in theory generalisable to any size of RD system. I had enough of an understanding of the maths to see what needed to be done, but being an experimental biologist actually doing the work was pretty slow progress. Coupled with the fact that I had just started a new postdoc and was therefore having to do a lot of this work in my free time, I sometimes thought we would never get to the end!
What next for you after this paper – I understand you’re now at the Crick?
AE: I have actually been at the Crick for a few years now in Caroline Hill’s lab – this piece of work with Jeremy took a lot longer to wrap up than we initially thought it would! I have been working on early zebrafish development, applying the kinds of computational and quantitative approaches I began to take while in Jeremy’s lab to look at the interactions between signalling pathways during early cell fate decisions in the embryo. I am now looking at options to start my own research group, although in the middle of a pandemic it feels as if this has become even more of a challenge than it normally would be.
Where will this story take the Green lab?
JG: I can see two ways forward with the rugae system. One would be to be more systematic about testing its robustness to perturbation. There’s room for both computational and experimental work to see, for example, whether perturbing two pathways simultaneously does what we predict. Another is to integrate what is really still a fairly minimal system with a transcriptomic picture of what the cells are doing. Have we really captured the key elements of the system, or are there others that we’ve just missed that a transcriptome of the periodicity would reveal?
Finally, let’s move outside the lab – what do you like to do in your spare time in London?
AE: I have 6 and 8 year old sons who have been occupying what spare time I have for the last few years! That said, they are now at an age where we do a lot of fun things together as a family, whether it be going on a bike ride, playing crazy golf or just having a lazy evening watching a film. At times when the work is full on, they certainly force me to switch off!
JG: To my own surprise, I’ve discovered, since working from home, how much I like to get exercise – even a bit of running which I used to hate! My wife and I have been cycling up the Thames in short stages, with a plan to reach the source in a few months, enjoying on the way some beautiful riverside scenery, some lovely little towns, and also a sense of being connected with the world, which is something that rivers give you, I think.


 (No Ratings Yet)
(No Ratings Yet)