The people behind the papers – Lisa Conrad, Steve Runser, Roman Vetter and Dagmar Iber
Posted by the Node Interviews, on 26 July 2021
This interview, the 97th in our series, was published in Development earlier this year.
Epithelial tubes perform crucial functions in various organs, providing routes for the transport of fluids and gases. A new paper in Development addresses the question of how epithelial tubes elongate during development, using a combination of mouse organ culture and mathematical modelling. To find out more about the work, we met four of its authors: PhD students Lisa Conrad and Steven Runser, senior scientist Roman Vetter, and their supervisor Dagmar Iber, Professor in the Department of Biosystems Science and Engineering at ETH Zurich.

Dagmar, can you give us your scientific biography and the questions your lab is trying to answer?
DI: I studied mathematics and biology, and did Masters degrees and PhDs in Cambridge and Oxford in each field. Since the very start, I have been interested in using mathematical modelling to uncover biological mechanisms. A Junior Research Fellowship at St. John’s College in Oxford gave me the freedom to pursue my own ideas and develop more precise, data-based models than the established conceptual models. Initially, I worked in immunology, but then switched to cell differentiation in bacteria, as the required quantitative data to build and validate models was available only for such simple organisms. These days, my group focuses on developmental mechanisms all the way up to human, and although we mostly collaborate with experimental groups, ETH allows me to also run a small wet lab to generate data and test ideas. The lab largely focuses on mouse lung and kidney development, but we maintain a rather broad interest in fundamental patterning mechanisms, and also collaborate with clinicians.
Roman, can you tell us a little about your research history and how you ended up working on developmental questions?
RV: I’m a computational physicist by training, and did my studies and PhD at ETH Zurich. With a few exceptions, my interest has always been in explaining nature’s wealth of emergent complex behaviour from simple, fundamental principles. During the earlier stages of my academic career, I found these problems mainly in the physical disciplines – from snow metamorphism to filament packing to crumpled shells and even fuel cell simulations. If you go through life with an open mind, open eyes and open ears, there’s an interesting question calling for a quantitative explanation virtually everywhere. My attention is drawn easiest by questions that combine geometrical shapes with mechanics and patterning. Lately, I have found such inspiration more and more in biology, where fundamental aspects of morphogenesis and development require taking new vantage points to advance further. I joined Dagmar’s group 2 years ago when she was looking for a senior biophysics modeller and I was looking to enter the field of computational biology to find new puzzles to solve – it was an instant match. I started as a postdoctoral researcher and recently became a senior scientist and lecturer in her group.
Lisa and Steve – how did you come to work in Dagmar’s lab and what drives your research today?
LC: I studied biology at the University of Freiburg and molecular medicine at Uppsala University. Developmental biology has fascinated me from the beginning; besides the interesting questions and methods in this field, the beauty of development is just so captivating! During a variety of lab projects, I noticed how much I enjoy projects that bring together expertise from different scientific backgrounds. By applying to the Life Science Zurich Graduate School, I found out about Dagmar’s group and got curious about their multidisciplinary approach to developmental biology. I started as the first PhD student with an experimental focus in the group’s then recently opened wet lab, eager to build on my experimental and research skills, while learning about new ways on how to tackle developmental questions from a different angle. It has been challenging to keep up with the aspects of the group’s research that are far from what I studied, but it’s also immensely rewarding when we can join forces to find new ways to better understand organ morphogenesis!
SR: I started my studies in cellular and molecular biology at the University of Strasbourg, but I quickly deviated towards more computational fields. I have always been interested in the design and application of mechanical simulations for the study of biological systems. To respond to this interest and to learn more about numerical approaches, I applied to Dagmar’s lab to do my master’s thesis. After completion of the thesis a little less than 2 years ago, Dagmar offered me the opportunity to continue in this field as a doctoral student. Since then, I have been developing and using different types of simulation models to study organ growth and morphogenesis.
How did you come to study tube elongation?
DI: The group had long been interested in lung and kidney branching morphogenesis. Although our ligand-receptor-based Turing mechanism could nicely predict where new branch points would form during lung and kidney branching morphogenesis, we noticed that the branch shapes that emerged in our simulations looked nothing like in the embryo because we were missing that bias in outgrowth that lets epithelial tubes of embryonic lungs and kidneys lengthen more than they widen.
Before this project, what mechanisms had been proposed for biased tube elongation?
DI: In lungs and kidneys, chemotactic movement towards a source of FGF10 or GDNF had long been noticed, and the extracellular matrix is thinner at the bud tips. In mammary glands, a constricting force had been proposed. In plants, hoop stress is a popular theory to explain their biased growth. We tested all those ideas, but none could explain the biased outgrowth of embryonic lung or kidney tubules. In fact, when checking the potential of hoop stress, we noticed that the lumen of the tubes is mostly very narrow, whereas the wall, the epithelium, is comparably thick. Although this is inconsistent with a role of hoop stress, it gave us the idea that fluid flow-induced shear stress may play a role.
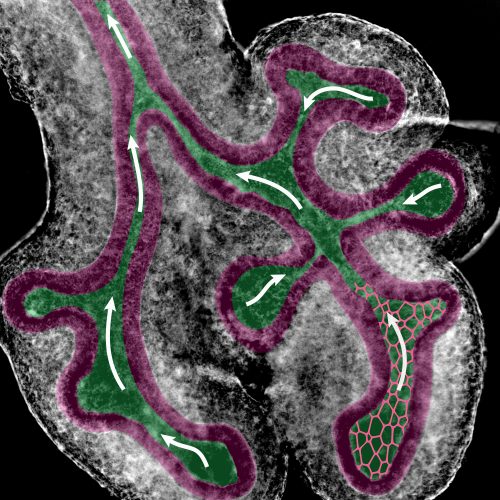
Can you give us the key results of the paper in a paragraph?
SR: The key result of the paper is that the observed bias in epithelial tube outgrowth, the accompanying bias in the apical cell shape and the resulting biased orientation of cell divisions, can be explained with fluid-flow driven shear stress. After having ruled out all the previously proposed mechanisms, Roman used a Finite Element method to demonstrate that the collapse of the tubes in itself was very unlikely to result in a bias in outgrowth. Instead, the narrow luminal region meant that a fluid flow in the epithelial tubes might cause a significant level of shear stress on the cell walls. To prove the existence of a flow, Lisa injected micro-beads in the lung lumens and observed their movements over time. We simulated the effect that a flow with the measured velocity would have on the cell walls of a similar lung tube geometry. The shear stress levels thus calculated were well within the range of what epithelial cells can sense. Shear stress is well known to result in the elongation of endothelial cells along the flow direction in blood vessels. I used a cell-based vertex model to investigate the impact of such an elongation on epithelial tissues. Once parameterized based on quantitative data, the model was able to recapitulate all the measured features of the lung and kidney tube epithelium. The bias in cell division orientations was well in line with what had been measured, and similarly the bias in outgrowth of the tissue matched with the experimental observations.
How do you think lung and kidney cells sense shear stress, and how is this sensing translated into biased growth at the tissue level?
DI: Epithelial cells can sense shear stress with their cilium. How they translate this into a change in cell shape, and how the extent of the cell shape change relates to the shear stress level is not known. Microfluidic experiments may help to resolve this.
Do you have any ideas for what causes tube collapse in early development?
RV: Indeed we do have some ideas, and we’re working toward testing possible theoretical explanations with detailed computer simulations by Steve, and toward validating them with experimental data from our wet lab, together with Lisa and other group members. Tube shape and collapse is an exciting topic of ongoing research in our group, and we’re looking forward to telling you more once we have conclusive answers.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
LC, SR: The project really picked up momentum when Harold Gómez, who is also shared first author, noticed, through careful examination of his beautiful lightsheet microscopy data, that the lumen is very narrow in many parts of the lung. But the most exciting moment was when we simulated the shear stress produced by the fluid flow, which we had finally succeeded in measuring experimentally, and realized that it was within the range cells can sense.
The most exciting moment was when we simulated the shear stress produced by the fluid flow.
And what about the flipside: any moments of frustration or despair?
DI: For me, certainly the first response by the referees. For years, I had asked experimental colleagues how to measure fluid flow in lung or kidney tubules and discussed strategies with my team. Yet, no one in my group felt they could pull this off. So, I decided to send the paper to Development, hoping that it would inspire some experimental lab to do that last experiment. After all, through a combination of (simple) experiments and simulations (that encompassed many different sophisticated techniques) we had excluded all previous proposals, suggested a new one, and provided convincing evidence for it. Yet, the referees would not have it. They not only insisted on that last experiment, but also saw little value in the paper as it was. I have seen it more than once that junior members got driven away from science by referee reports that had failed to recognise the value of their work. But not so Lisa: realising that those fluid-flow measurements were the make-or-break, she decided to just give it a try – despite all COVID restrictions. She remembered that Renato Paro had left an injector to our department when retiring, which no one used. She got it to work with the help of his former technician – and demonstrated fluid flow at the required level in the developing lungs.
LC: For me, the biggest source of frustration is when an experiment fails due to technical issues and the tissue samples that I harvested from an animal go to waste. Regarding the referee reports, Dagmar encouraged us to take on the challenge, and celebrating the small successes along the way kept spirits up! In the end, confirming our proposed mechanism was a really good experience.
How has your research been affected by the COVID-19 pandemic?
LC: Last year in March, ETH went into lockdown to help slow down the spread of the virus and our lab was closed for about 6 weeks. The reopening was done in several phases, so we had to work in shifts for a while and it took some time to start up experiments again. Working in shared facilities is still restricted, but most of the time we can figure that out by communicating with colleagues and working around each other’s schedules. In general, everything needs a bit more planning now. For the paper revisions, we had to set up a new experiment in the lab while COVID-19 restrictions were still in place, which of course provided an extra challenge. Having a colleague take a look at your (failed) experiment usually speeds up optimization and trouble-shooting and there are fewer opportunities for spontaneous exchange with other groups. Despite the additional work and stress brought by the pandemic, I feel like everyone at BSSE is supporting each other; especially Makiko Seimiya and Tom Lummen (BSSE Single Cell Facility), who have been awesome in sharing their advice and equipment, and helping me with many tries at the spinning disk confocal, which was a new microscopy system for me.
DI, RV, SR: As theoreticians, we have been forced into a home office for more than a year now. We miss the personal interactions, but remote work is otherwise straightforward for us.
What next for you two after this paper?
LC: I am currently finishing up a project that compares branching morphogenesis in lungs and kidneys. In a collaboration with Roman and other team members, we’re also looking at tube formation, where I’m using nephrogenesis in kidney organoids as a model.
SR: The 2D vertex model used in this paper offers many possibilities for the study of tightly packed cell sheets. However, numerous developmental events can only correctly be represented in three dimensions. With the advent of high performance computing, new computational frameworks representing cells more realistically in 3D can now be developed. I am currently developing such a model specifically designed for the simulation of epithelial tissues.
Where will this story take the Iber lab?
DI: The mechanism that defines the aspect ratio of tubes is one important puzzle piece to explain how complex organs are shaped and how organ-specific differences arise. There are many other fundamental questions concerning tubulogenesis, epithelial organisation, the physics of budding, the role of the mesenchyme and the final reorganisation of the lung epithelial tree into a fractal-like architecture. As a group, we are interested more generally in self-organisation during development.
Finally, let’s move outside the lab – what do you like to do in your spare time in Basel?
LC: I picked up diving a few years back and it’s so much fun! For a local spot, I would recommend the Bodensee; it’s really amazing to dive into a whole new world ‘at home’, now that travelling hasn’t really been an option. Basel has lots of relaxing greenery and I often go for walks along the Rhine. Since the start of the pandemic, I have also rediscovered the fun in crafting and caring for plants on my balcony.
SR: I enjoy a lot of different activities, which range from reading historical novels to playing football.
RV: Spare time comes and goes in phases. There are more things I’d enjoy doing than I could possibly fit into a full day – cycling across the country is one of many.
DI: I enjoy the Swiss mountains, swimming in the Rhine, the tennis court in front of my house – and the preparation for SoLa, the yearly running relay race, which we participate in with the entire group.


 (No Ratings Yet)
(No Ratings Yet)