The people behind the papers – Mingxi Deng and Yan Yan
Posted by the Node Interviews, on 7 November 2019
This interview, the 70th in our series, was recently published in Development.
Drosophila wing discs are epithelial sac-like organs and a powerful model for investigating the link between proliferation and patterning. Of particular interest is the question of how single cells in the disc integrate information regarding position and growth control, as morphogens that pattern an axis can also regulate cell division. A new Techniques and Resources article in Development reports the application of single cell sequencing technologies to dissociated discs in an effort to understand these problems. We caught up with first author Mingxi Deng and his supervisor Yan Yan, Assistant Professor at The Hong Kong University of Science and Technology (HKUST), to hear more about the story.

Yan, can you give us your scientific biography and the questions your lab is trying to answer?
YY My lab is primarily interested in organ size control, in particular, the roles of cell structural components such as apicobasal polarity proteins and cytoskeletal proteins in this process. When I was a graduate student with Prof. Trudi Schupbach at Princeton University, New Jersey, USA, I performed a genetic screen for mutants affecting Drosophila follicle cell epithelial morphogenesis and proliferation. I then got my postdoc training with Prof. Chris Doe at the University of Oregon, USA, where I learned how Drosophila embryonic neuroblasts lose their apical domains and emerge from neuroepithelia. These experiences were important for me to learn that cell polarity and, more broadly, cell structural proteins, are important for organ size and shape. Another important thing I learned from graduate school is the power of quantification, which influences how we approach questions now in the lab.
Mingxi, how did you come to work in the Yan lab, and what drives your research today?
MD When I was looking for a postgraduate student position at HKUST I found Prof. Yan’s research interesting in combining the power of Drosophila genetics with quantitative biology methods. Her lab also has a good reputation for being supportive to students, so I decided to join. I have always wanted to become a scientist and I get excited when I encounter new problems and need to find a way to solve them.
What was the drive behind doing a single cell analysis of the disc, and how easy was it to set the system up?
MD & YY It started with our study of how scribble (scrib) mutant tumours – which show disrupted tissue architecture – change over time. We found that they showed a high degree of plasticity, and suspected that they might be more heterogeneous than previously assumed. For this we needed to understand how much of the cell heterogeneity in the scrib tumours comes from heterogeneity already existing in wild-type wing discs. That was the starting point of this analysis.
We were lucky to have the help we needed for this study. Prof. Ting Xie from the Stowers Institute for Medical Research, Missouri, USA, happened to visit my university at the time. I went to talk with him and he kindly shared his unpublished fly single cell dissociation protocol. It is also very helpful to have colleagues Jiguang Wang and Hao Ge with whom to discuss methods: their expertise in bioinformatics and mathematics ensures that we are analysing data correctly and robustly. In addition, the single cell community has been very good in providing open-access analytical tools with user-friendly tutorials.
Can you give us the most surprising finding from your paper?
MD & YY The most surprising finding is that pattern formation partially persisted in the scrib mutant tumours. This is surprising because the morphogens important for pattern formation need to properly spread in space, and it suggests that further studies are needed to understand why particular pattern formation processes are robust against loss of tissue architecture.
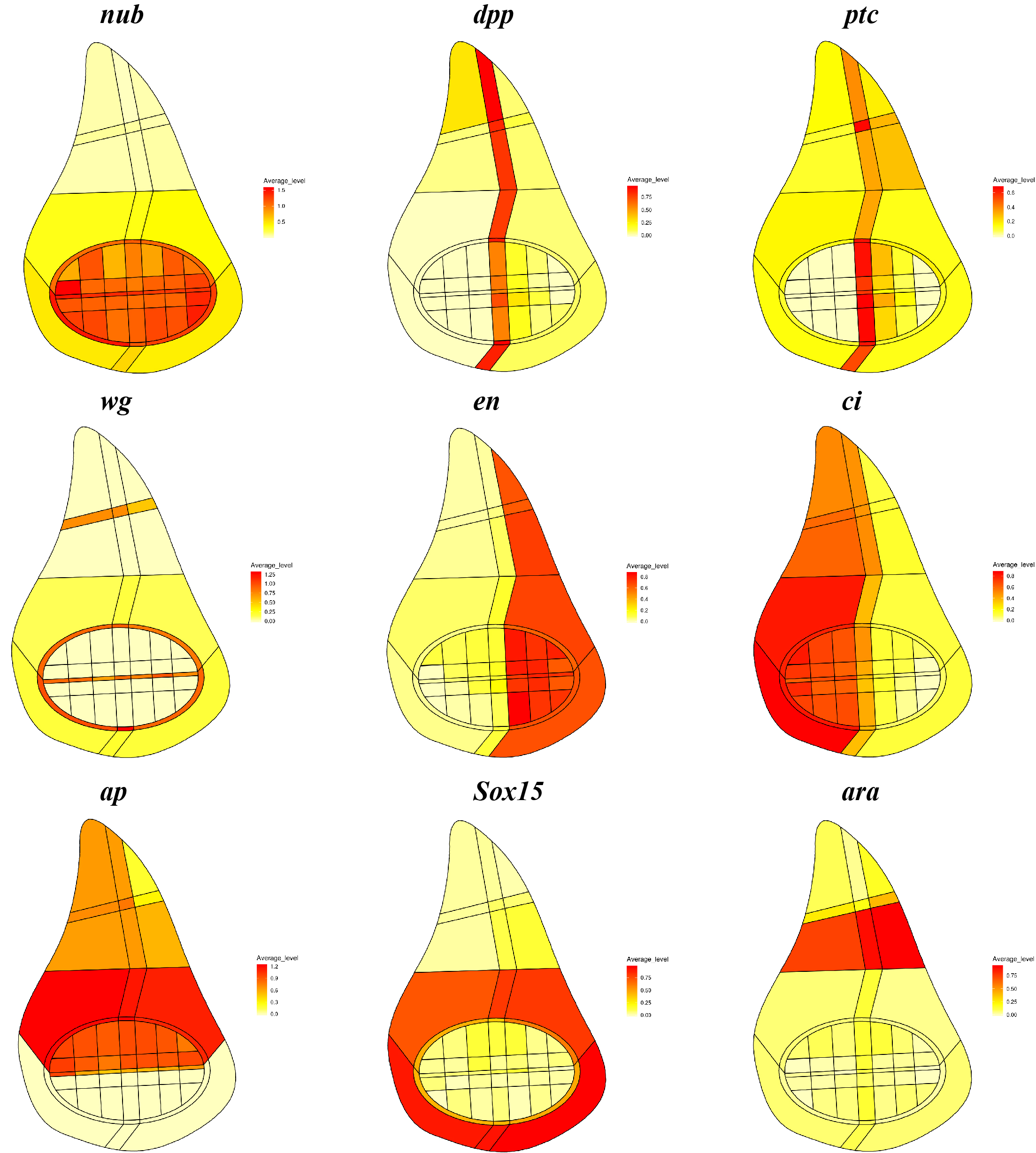
What does your scrib tumour model analysis tell you about the link between patterning and growth control in the disc?
MD & YY This is a very good question, but we still do not understand the link between patterning and growth control, although we are able to make more quantitative observations from these data. It was previously shown, with very clear genetic evidence, that patterning factors such as dpp are needed to ensure proper proliferation and growth in the disc. Previous experiments have also shown that cell proliferation and growth are more or less uniform throughout the whole disc. Now, we provide another line of evidence that proliferation and growth states do not appear to be particularly biased in sub-regions marked by any single patterning gene, in both wild-type and scrib mutant discs. Our data also suggest that a well-defined distribution of proliferation and growth states exists in discs and this distribution is severely disrupted in the scrib mutants. Interestingly, the temporal scrib mutant data suggested a positive correlation between formation of correct patterns and a distribution of proliferation and growth states closer to wild type.
Your single cell datasets are available to explore on a database: what questions do you think this database will be particularly useful for addressing?
MD & YY Wing discs have been a very good system to study pattern formation, organ size control and regeneration. I hope that our database can provide a good reference point for the community interested in these questions. For example, for researchers interested in how wing disc cells respond to injury during regeneration processes, they would be able to compare the identity of their cells of interest with the wild-type imaginal disc cells in our database.
I hope that our database can provide a good reference point for the community
When doing the research, did you have any particular result or eureka moment that has stuck with you?
MD After I assigned the disc cells correctly to the pouch/hinge region, I was very happy to see that fine patterning processes are well represented in our single cell data. This gave me a sense of connection and also deep respect for the classical works on pattern formation, which I had previously only learned from textbooks.
And what about the flipside: any moments of frustration or despair?
MD I started as the only student working on computation in our lab and needed to learn everything from zero. My lab mates are all excellent experimentalists but they cannot help me with computational problems. It took a while to grow out of the loneliness but I have become more confident now.
So what next for you after this paper?
MD I have just finished my second year as a postgraduate student. I am now pursuing a few quantitative biology projects for which we already have data and a priority for me is to further sharpen my computational and mathematical skills. Hopefully, I can share new exciting stories in a few years when I graduate with my PhD.
Where will this work take the Yan lab?
YY The scrib mutant cells are very interesting, because when they are generated as mosaic clones in the wing discs they behave very differently and undergo cell death through a cell competition process. Building upon this work, we are now trying to better understand the scrib mutant clonal cells, and how different signalling activities contribute to their cell plasticity at the single cell level and eventually alter their growth outcome.
Finally, let’s move outside the lab – what do you like to do in your spare time in Hong Kong?
YY: I have a 4-year-old son and I am expecting another baby in December, so my activities outside the lab revolve around parenting. I find the parenting experience extremely helpful in that I am much more patient with students now than before.
MD: Hong Kong is a surprisingly great place for outdoor activities like hiking and sailing, which I like. I also like to play soccer and computer games.


 (No Ratings Yet)
(No Ratings Yet)