Applying tandem timers to measure signalling and gene expression dynamics in developing embryos
Posted by Joseph Barry, on 11 January 2016
The signalling systems that conduct the orchestra of embryonic development are fantastically complex and dynamic. We owe much of our knowledge of in vivo signalling dynamics to advances in microscopy and protein tagging with fluorescent reporters that have allowed visualization of signalling proteins. Looking forward, however, it is clear that simply analyzing the localization patterns of proteins is only the first step to fully understand signalling processes. Of course, a protein’s presence at a particular time or place in the embryo does not tell us if it is actively signalling or lying dormant until its activity is required. Therefore, generic methods to distinguish actively signalling proteins from passive proteins will be tremendously useful advances for researchers interested in developmental processes.
A few years ago, Darren Gilmour at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, realized that since many signalling events lead to the degradation, stabilization, or relocalization of participating proteins, methods for detecting changes in protein turnover could provide powerful proxies for in vivo signalling activity. Gilmour and his graduate student Erika Donà therefore took advantage of a dual-color genetically encoded fluorescent reporter called a “tandem timer”, newly developed by Michael Knop in the next-door lab at the EMBL. Tandem timers report on the age of protein populations through the relative fluorescence of adjacent slow- and fast-maturing fluorophores (Khmelinskii et al. Nat. Biotechnol 2012). In collaboration with Knop and his postdoc Anton Khmelinskii, Gilmour and Donà tagged the chemokine receptor Cxcr4b with a tandem timer with a view to observing Cxcl12a chemokine signalling activity across a migrating tissue, the zebrafish posterior lateral line primordium. They wagered that this strategy was likely to inform on chemokine signalling activity since Cxcr4b receptors undergo rapid internalization and degradation upon ligand-mediated activation. Higher Cxcl12a levels would increase Cxcr4b internalization rate and this should be reflected by a decrease in the receptor’s population age (increased protein turnover). Gilmour and Donà also understood the potential long-term implications of their attempt; a successful proof-of-principle study might lead to the adoption of the tandem timer as a generic tool for the detection of signalling activities.
At the time I was a postdoctoral researcher in Wolfgang Huber’s group at EMBL, and having collaborated with Knop and Khmelinskii on the original tandem timer paper, I was happy to get involved in this first project using tandem timers in embryos. Using image processing and analysis tools from R and Bioconductor software, I measured tandem timer signalling readouts from 3D images of migrating tissues, and used a mathematical model of timer behaviour to interpret the results. The model describes the maturation kinetics of the two fluorophores on the timer, as well as production and degradation rates of the tagged protein. What surprised and delighted me throughout my collaboration with Gilmour and Donà, was the extent to which quantitative analysis and modelling was able to feed back on subsequent experimental design, and positively influence later results. Ultimately our tandem timer observations, along with other data, allowed us to demonstrate that a self-generated chemokine gradient guides the migration of the zebrafish posterior lateral line primordium (Donà et al. Nature 2013).
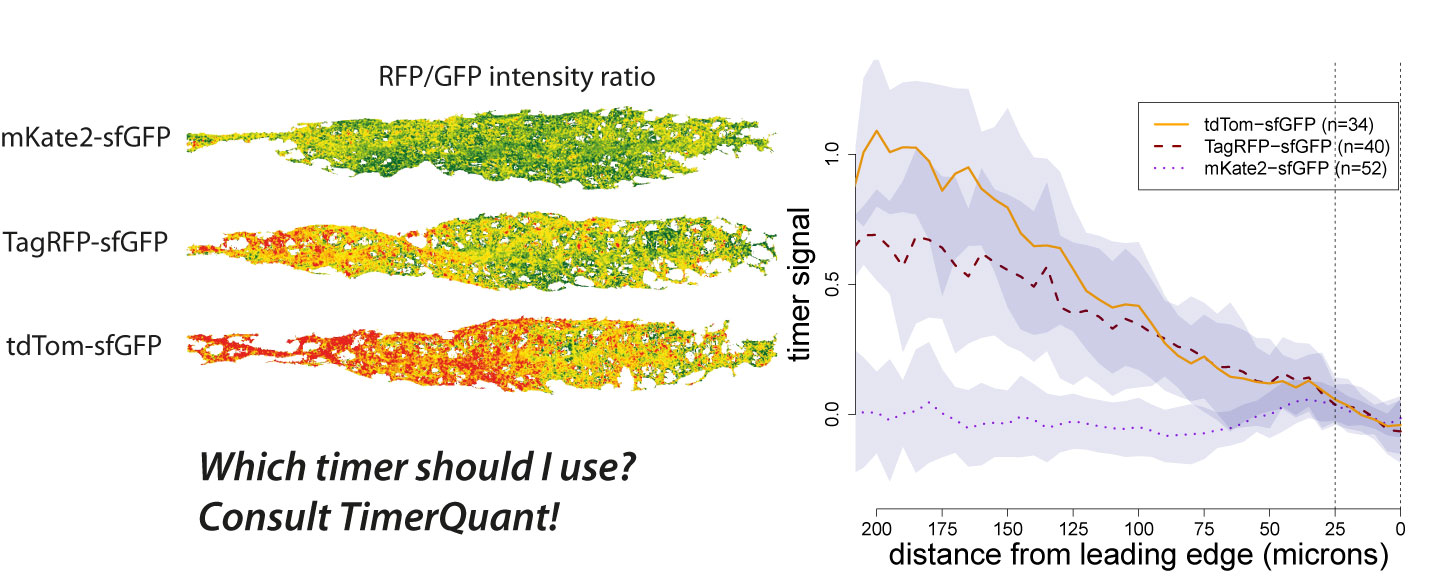
Tandem timers can be constructed with virtually any combination of spectrally separated fluorescent proteins with different maturation rates. Therefore, the first question an experimentalist typically asks when using tandem timers is: which fluorophores should I choose? We have been contacted by many groups asking exactly this question. As our ability to design timers and interpret timer data has benefited greatly from the use of modelling, we decided to write a paper outlining how our modelling tools could be used by other researchers interested in tandem timer research. To make these tools easily accessible we developed an interactive web application, TimerQuant, and made all our code available through the open-source software platform Bioconductor. For our new paper, which is now published in the journal Development (Barry et al. Development 2016), we systematically investigated the effect of relevant experimental parameters on timer signal, a measure of how good a particular timer is at detecting differences between two protein half-lives. To validate the predictions of our model, we reinvestigated the Cxcr4b signalling gradient identified in our original study using three tandem timers that had the same fast-maturing fluorophore (sfGFP), but different slow-maturing fluorophores (mKate2, mCherry, TagRFP). We found that timer signal decreased as the maturation times of the slow- and fast-maturing fluorphores became more similar. Readouts became noisier as protein abundance decreased. Timer signal increased with the maturation time of the slow-maturing flurophore, albeit at the expense of noisier readouts. While these conclusions might already be expected from a more qualitative reasoning, the quantitative model also led to some unexpected and intriguing findings. For example, Förster resonance energy transfer (FRET) between the fast- and slow-maturing fluorophores actually increased timer signal, a prediction that was borne out in our experiments, and which may be an important consideration for future timer designs.
In the initial model protein production (expression) and degradation rates were kept constant. Given that in developmental contexts these are likely to change over time, we decided to explore the effect of dynamics on timer ratio profiles. We modelled constant increases and decreases of protein expression and degradation rates as well as sudden bursts of expression and degradation, and looked at model solutions over time. The simulation results showed timer ratio profiles over time producing clear, characteristic responses to expression and degradation dynamics. We were surprised by the extent to which expression and degradation responses were distinguishable from one another. These additions to the model further help in the interpretation of timer ratio results in non-steady state conditions, which is often the situation when studying dynamic developmental processes. Moreover, as degradation rate could be experimentally fixed, for example by using non-degradable (or slowly degradable) versions of tandem timers, these model predictions open up the exciting possibility of using tandem timers to observe gene expression dynamics with time-lapse microscopy.
Explore Further:
Interactive TimerQuant software applications including those showing timer responses to dynamically changing gene expression can be used online at http://chronos.embl.de/TimerQuant/ or offline through the TimerQuant software package on the Bioconductor website.
Papers Referenced:
1. Barry JD, Donà E, Gilmour D and Huber W, TimerQuant: A modelling approach to tandem fluorescent timer design and data interpretation for measuring protein turnover in embryos, Development 143(1), 2016
2. Donà E, Barry JD, Valentin G, Quirin C, Khmelinskii A, Kunze A, Durdu S, Newton LR, Fernandez-Minan A, Huber W, Knop M and Gilmour D, Directional tissue migration through a self-generated chemokine gradient, Nature 503(7475), 2013
3. Khmelinskii A, Keller PJ, Bartosik A, Meurer M, Barry JD, Mardin BR, Kaufmann A, Trautmann S, Wachsmuth M, Pereira G, Huber W, Schiebel E and Knop M, Tandem fluorescent protein timers for in vivo analysis of protein dynamics, Nature Biotechnology 30(7), 2012


 (4 votes)
(4 votes)