How to build and evolve a nervous system
Posted by VeronicaHinman, on 20 November 2016
Author Summary of “A gene regulatory network for apical organ neurogenesis and its spatial control in sea star embryos”.
Alys M. Cheatle Jarvela, Kristen A. Yankura, Veronica F. Hinman
Development 2016 143: 4214-4223; doi: 10.1242/dev.134999
Very similar cell types are found across the animal tree of life. Much of animal diversity, therefore, arises not from the formation of new cells, but from the evolution of the developmental control of the numbers and patterns of orthologous cell types. Neural cells types are a particularly interesting example of this phenomenon. Almost all animals make serotonergic neurons, and for example, these neurons form at the anterior pole of many invertebrate larval types and in the anterior nervous systems of vertebrates. The numbers, distribution, and relative locations of serotonergic neurons are however quite varied. Thus animal gene regulatory networks (GRNs) must function to make this very conserved neural type, but be able to evolve the patterning and numbers of these cells.
We have used the sea star, Patiria miniata, as a model system to ask how serotonergic neurons are made, and how they are positioned, so that we can understand this phenomenon. Sea stars are representatives of one class (Asteriodea) of the Phylum Echinodermata, which also include sea urchins (Cl. Echinoidea), and sea cucumbers (Cl. Holothuroidea). The sea star larvae, like the larvae of many invertebrate marine organisms has just a small number of serotonergic cells and these are readily visualized by immuno-localization using an antibody against serotonin (Fig 1).
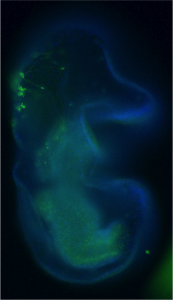
Figure 1. Immuno-localization of serotonergic neurons in the sea star bipinnaria larva. Lateral view, mouth is to the right, anterior is up, and dorsal ganglion is top left.
From previous work (Yankura et al., 2013) it was known that there are many soxc expressing cells scattered throughout the ectoderm in the two-day old gastrula that will form, not only the serotonergic neurons, but also other types of neurons in the later three to four-day larva. However, by the time neurons differentiate into various neural subtypes, the expression of soxc is extinguished. These cells were therefore labeled with a stable GFP construct to show that cells that originally express soxc do indeed end up forming serotonergic neurons (as well as other neural cell types) (Fig 2). Thus soxc+ cells in sea stars are neural progenitors, and these progenitors are distributed broadly across the ectoderm. The progression of these progenitors to differentiated neurons occurs through a series of asymmetric divisions (Fig 2). In particular one of the soxc+ daughters will express the LIM homeodomain transcription factor lhx2/9, and in turn lhx2/9 expressing cells will undergo both symmetric and asymmetric division, the asymmetric division producing a finally differentiated serotonergic neuron. Thus the lhx2/9 cells represent a proliferating, more restricted pool of serotonergic progenitors.
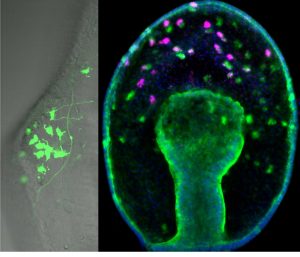
Figure 2. A stable GFP construct, driven by the soxc cis-regulatory region, is localised to neurons of the dorsal ganglion in the larval stage on left image. Right image: soxc expression cells (green) are found scattered throughout the ectoderm in the earlier gastrula. One of the soxc+ daughter cells expresses lhx2/9 (pink) in only the more anterior territory.
Next, the spatial domains in which these asymmetric divisions were occurring was examined. It has been known for many years that many animals, including the sea star larva (Yankura et al., 2010), share a remarkably conserved patterning along the AP axis. Genes with roles in patterning the anterior-most nervous system in vertebrates, for example, are also expressed in the most anterior regions of the larvae, and genes involved with patterning midbrain-hindbrain region are expressed at the posterior boundary of the ectoderm. This new work now shows that these AP domains control neural progression. That is, soxc+ cells divide to produce two soxc+ cells in the posterior zone, asymmetrically divide to produce an lhx2/9+ sister in the intermediate zone; lhx2/9+ cells will also divide symmetrically in this mid-zone, and will exit proliferation and become differentiated neuron in only the anterior-most, foxq2 expressing zone. These AP domains, therefore, establish neural proliferation zones.
Altering the size of the AP zones caused predicted changes in neural proliferation, and therefore changed the final numbers of serotonergic neurons. This shows therefore that a function of these highly conserved territories, in sea stars at least, is to regulate neural cell type progression.
This new work, therefore shows that neural progenitors form throughout the ectoderm without regard to patterning in the sea star, and that the GRN that establishes domains along the AP axis, controls neural progression. We predict that fairly simple evolutionary changes to this patterning GRN could change the size of these territories, and hence the numbers and distributions neural cell types.
REFERENCES
Yankura, K. A., Martik, M. L., Jennings, C. K. and Hinman, V. F. (2010). Uncoupling of complex regulatory patterning during evolution of larval development in echinoderms. BMC Biol 8, 143.
Yankura, K. A., Koechlein, C. S., Cryan, A. F., Cheatle, A. and Hinman, V. F. (2013). Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc Natl Acad Sci U S A 110, 8591–8596.


 (2 votes)
(2 votes)