Dating with cells – finding the right match
Posted by dbsste, on 23 August 2018
It’s an age-old mystery of the heart: do opposites attract, or will like do better with like? We can now answer this pressing question, at least for Drosophila cardioblasts: cells prefer to ‘swipe right’ on a shared transcriptional profile, but the resulting relationships are stronger if there are some unattractive alternatives around to remind them to love the one they’re with.
To put that in more scientific terms, in order to build complex structures that perform versatile functions, biological systems need to be able to specific and precise cell-cell connections. Yet the question of how cells find the right partners as organs form generally remains poorly understood. In our recently published work (Zhang et al., 2018), our team has uncovered some of the secrets of how cells make the perfect match.
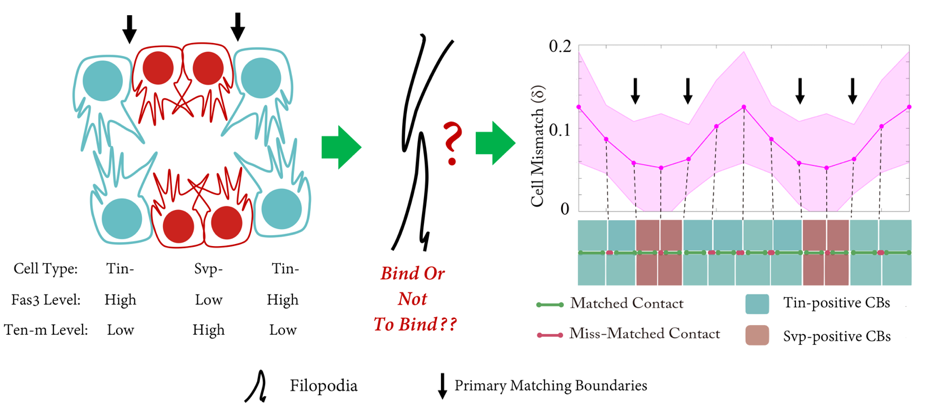
The Drosophila heart tube is constructed from two parallel lines of connected cardioblasts in the developing mesoderm, initially separated by over 100mm. The cardioblasts migrate together and create the heart tube during stage 16. The cardioblasts have distinct subtypes: Tinman-positive cells that form the heart lumen and valves; and Seven-up-positive cells that form the ostia. The Drosophila cardiogenesis ‘dating algorithm’ has Tinman and Seven-up-positive cardioblasts lined up in a repeating 4-2 pattern (Figure 1). As the contralaterally opposing lines of cardioblasts come together, they only match up with cells of the same type (Figure 1). We set out to investigate: how does this happen, and why?
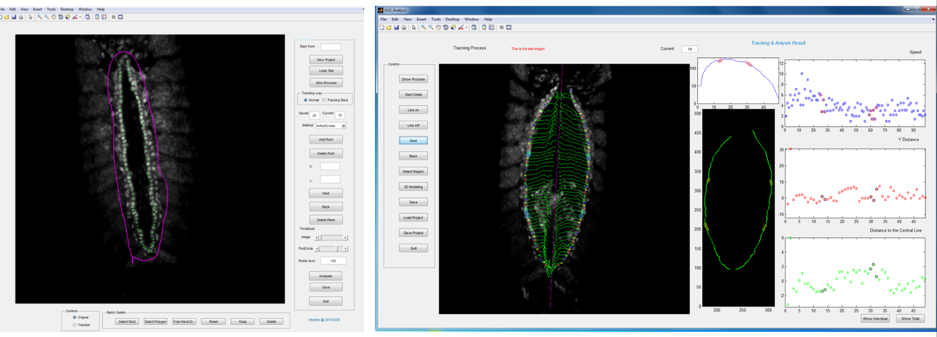
Primary ‘matchmaker’ Shaobo Zhang, as part of an undergraduate research project, had the job of figuring this out. He was working under tough conditions: less than a year old, the lab had few reagents and no fly room. Nevertheless he got his hands on a Hand::GFP line, which expresses a marker for cardioblasts in the Drosophila embryo, and developed cell tracking software to see how the cells migrate during heart formation (Figure 2). The project proved compelling enough to persuade Shaobo to join the lab (https://mbi.nus.edu.sg/timothy-saunders/) for his PhD studies, seemingly undaunted that the PI was a theoretical physicist. (It probably helped that by then we had a mini fly room – albeit with three levels of security to stop any flies escaping, Figure 3).
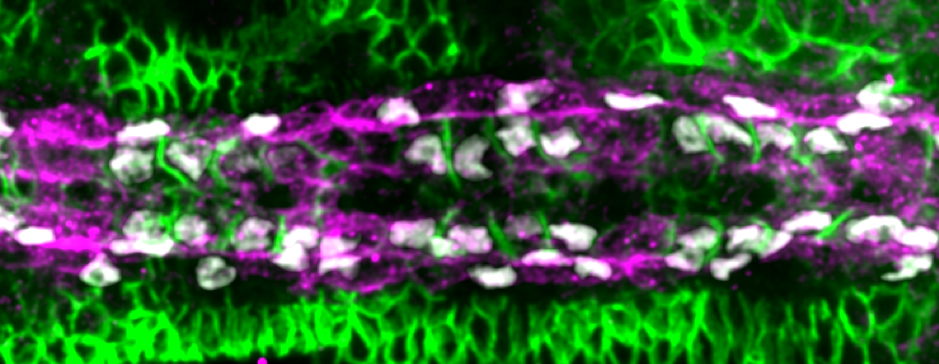
Shaobo noticed that – rather like competitors on Love Island – cardioblasts could change the partner they coupled up with quite abruptly. As the team looked more carefully, we realised that cells were sampling their local environment using filopodia protrusions. However, not all cells were equally good at finding their partners precisely. The best matched cells were at the boundary between the Tinman- and Seven Up- positive cardioblasts.

But what factors helped cells decide which of the opposing cells was the right one for them? We notice that the filopodia forming strong connections were generally from the Tinman-positive cells. In contrast, the filopodia interactions between Seven Up-positive cells were distinctly unromantic, and rather more like Neymar and Ronaldo having a fight – a lot of arm-waving and drama, but no meaningful contact.
Shaobo reasoned that cell-cell adhesion molecules may be differentially expressed in cardioblasts to facilitate the selective filopodia adhesion. At this point he turned to the literature, as neurobiologists have been investigating neuronal cell matching for years and a range of known “matching” molecules are known, though the dynamic mechanisms through which they act are not fully understood.
Performing a mini-screen of these targets, we found that Fasciclin III (Fas3) stood out. Fas3 is a homophilic adhesion molecule (Figure 4), which was more highly expressed in the Tinman-positive cardioblasts. Perturbing the expression pattern of Fas3 resulted in perturbations of their filopodia binding activities, leading to increased cell mismatch.
At this point, it looked like we had found the molecule driving cell matching (a bit like alcohol at a student party). However, when we looked at fas3-/- mutants we noticed only a small defect in cell matching (unbelievably, dating can also occur sober). Returning to our screen results, we noticed that the adhesion molecule Ten-m (also known as Teneurin-m or Tenascin-m – sometimes naming conventions really need to be sorted out) was upregulated in Seven-up positive cells (Figure 4). After some painful crosses, Shaobo produced the double mutant of Fas3 and Ten-m, which, thankfully, had a significant matching phenotype. Therefore, it appears that heart cells use two (partially redundant) adhesion molecules to ensure they find the right partner.
But how is this differential expression pattern genetically regulated? To answer this, we turned to Dr David Garfield at Humboldt University in Berlin (https://www.garfieldlab.org/). By looking at putative enhancers specific to mesodermal and neuronal tissue, he identified potential tissue-specific enhancers for Fas3. Shaobo made reporter lines to test whether these distinct regions correspond to the specific expression patterns for Fas3. Thankfully they did, with specific expression in cardioblasts (with differential expression in distinct cell types) and neurons. So – for cardioblasts anyway – the odds of pair-bonding are partly a matter of genetic destiny.
This project was a lot of fun as we explored how both mechanical and genetic mechanisms interplayed to regulate the precise cell matching and help forming the properly structured heart. Given the conservation of many of the genes involved in early heart formation, we are hopeful that this will have relevance to vertebrate systems. More interestingly, this shows a simple but potentially general dynamic mechanisms of constructing specific cell-cell connections in biological systems. We’ll keep you posted if we crack the human relationship code too.
Reference
Zhang, S. et al. (2018) ‘Selective Filopodia Adhesion Ensures Robust Cell Matching in the Drosophila Heart’, Developmental Cell, 46(2), p. 189–203.e4. doi: 10.1016/j.devcel.2018.06.015.
https://www.cell.com/developmental-cell/abstract/S1534-5807(18)30500-8


 (4 votes)
(4 votes)