A fine balance: tuning adult neurogenesis in the freshwater planarian
Posted by kocurrie, on 12 January 2017
Comment on “Neuronal sources of hedgehog modulate neurogenesis in the adult planarian brain”, eLife 2016;5:e19735.
Currie et al., eLife (2016)
Ko W. Currie, Cellular Neurobiology, Institut de recherches cliniques de Montreal (IRCM), Montreal, QC
Alyssa M. Molinaro, Hospital for Sick Children, Toronto, ON
Bret J. Pearson, Hospital for Sick Children, Toronto, ON
To say that biologists have been studying freshwater planarians for a long time is a huge understatement, with one of the earliest known description of planarian regeneration dating back to 1766 (Pallas 1766). Many early naturalists and embryologists marvelled at the planarian’s ability to completely regenerate from any injury, even re-constructing an entirely new body plan from tiny tissue fragments. Even the famed Drosophila geneticist Thomas Hunt Morgan, enjoyed a brief stint experimenting with these flatworms, before moving on to the problem of genetic inheritance (Morgan 1898). Despite this earlier interest, planarians largely fell out of fashion as a model system for developmental biology for roughly an entire century.
I joined Bret’s lab in the Fall of 2010, at a time when planarian research had just enjoyed a dramatic 10-year resurgence. We were now able to specifically visualize the planarian stem cells (also known as “neoblasts”) that granted these flatworms their regenerative powers, and had uncovered some of the genes that regulated their proliferation and differentiation into the various somatic cells of the mature organ systems (Reddien et al., 2005). The genome of the most commonly used lab species, Schmidtea mediterranea, had recently been sequenced (Robb et al., 2008), and many of the fundamental cell signaling pathways, including the Hedgehog pathway, had been assessed for functions during large-scale regeneration events (Gurley et al., 2008; Reddien et al., 2007; Rink et al., 2009). However, significantly less attention had been focused on the planarian nervous system, which must be precisely re-constructed following decapitation, so that the new organ grows to the correct size in proportion with the rest of the body and that all of the mature neurons are regenerated in the proper cellular ratios, thereby bringing about full recovery of brain function.
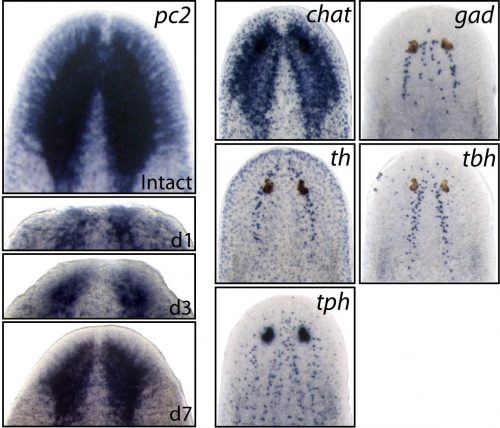
Earlier studies demonstrated the surprising molecular complexity of the planarian nervous system, but the specific genes and signaling mechanisms that regulated its construction were still largely unknown. Therefore, we set our sights on some very basic questions; 1) Within the large pool of planarian neoblasts, are there dedicated neural stem cells that are required for brain regeneration? 2) How are specific neural cell types specified/differentiated from upstream stem cells? 3) What extrinsic cell signals control the rate of neurogenesis during regeneration and in homeostatic (uninjured) conditions?
With these broad goals in mind, we did what most people do when you don’t have a concrete starting point, we did a screen. Taking advantage of the fact that planarians are susceptible to RNAi gene knockdown through feeding (a mixture of beef liver paste and E. coli that are forced to express double-stranded RNA for your gene of interest), it was possible to perform a high-throughput RNAi screen. We identified over one hundred planarian homologs of well-established neuronal regulatory genes, including many homeobox transcription factors, and proceeded to knock each one down, testing for possible functions in the planarian nervous system.
Since we were knocking down homologs of canonical neural stem cell regulators such as Pax6 and Sox2, we were really hoping that following decapitation, some of these RNAi-treated flatworms would exhibit major defects in brain regeneration. Alas, as is often the case in science, we never got such striking results. However, what we did notice, was that a fair number of RNAi-treated worms (even without decapitation) exhibited an array of strange/aberrant behaviours (including paralysis, and some very odd involuntary muscular contractions), that we thought could be caused by neuronal dysfunction within the intact nervous system. Therefore, we decided to focus on the transcription factors which yielded behavioural defects after RNAi-knockdown, examining them for neuronal specification defects.
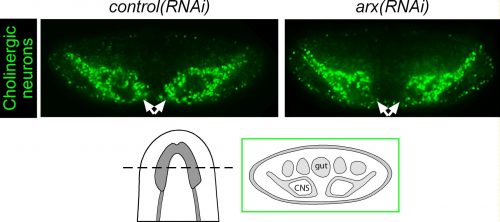
Among these genes causing strange animal behaviours were two homologs of homeobox transcription factors, nkx2.1 and arx. In particular, knockdown of nkx2.1, caused a very striking muscular contraction defect seen above (colloquially called the “cobra” phenotype in our lab), resulting in the animals constantly rearing back their heads so that they swam with their head perpendicular with the rest of the body, similar to a cobra snake. When we examined the brains of planarians after RNAi against nkx2.1 and arx, we found that these worms were missing a significant number of neurons belonging to the cholinergic, GABAergic and octopaminergic neural subtypes, specifically in the medial-most region of the brain (see image below), implicating these two transcription factors in the specification of these neuronal populations. Interestingly, we also found planarian stem cells immediately adjacent to this brain region that expressed nkx2.1 and arx, leading us to speculate that within this microenvironment (see image below), there may be communication between the mature neurons and their upstream stem cells to regulate their own production.
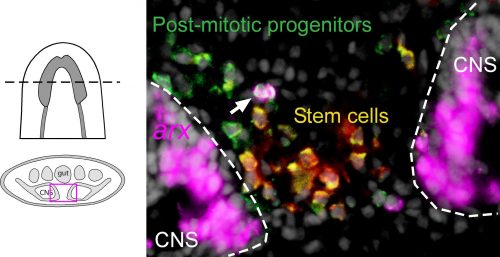
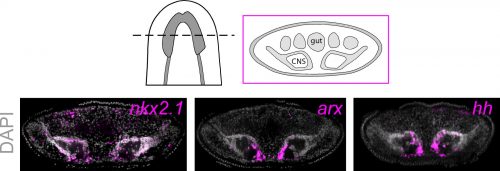
To investigate what this signal might be, we decided to look at the Hedgehog signaling pathway. It was already known that the planarian hedgehog (hh) ligand was expressed in the medial regions of the CNS (Rink et al., 2009). But we showed more specifically that the hh signaling molecule was actually expressed in the very same medially-located neuron subtypes as nkx2.1 and arx. In addition, we found that stem cells located right next to the brain expressed the Hedgehog receptor patched, as well as the downstream effector genes, smoothened and gli-1, demonstrating that planarian stem cells were likely capable of receiving and responding to hh signals coming from the mature brain.
Next, we had to actually test whether hh signaling had any functional consequence on neurogenesis levels. For all of the interesting biology and techniques that are now available within the planarian research community, we still don’t have true genetic tools such as transgenics to perform gene knockout or overexpression experiments in a targeted cell-specific manner. Instead, we had to rely on organism-wide gene knockdown experiments to globally decrease or increase hh signaling, which admittedly I was a little nervous about, due to the wide range of roles that the hh pathway plays in animal development. Luckily, we found that when we knocked down the planarian hh ligand (decreased signaling activity), this resulted in the production of fewer neural progenitor cells as well as new mature cholinergic neurons, while having little to no effect on other tissue lineages (skin, gut & pharynx).
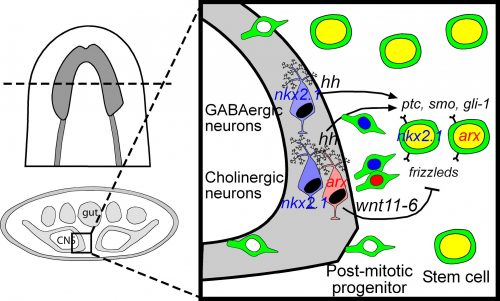
We were initially a little confused by the result, since one would expect that in an intact “steady-state” brain, any signals coming from these mature neurons would actively repress the production of new neurons, whereas brain injury or the removal of nervous tissue would promote stem cells to increase their neuronal output. Instead, our results indicate the opposite, where hh signals coming from medial neurons are actually required to promote adult neurogenesis. It is possible that hh acts to counter and balance out the effects of local Wnt signaling, which has been shown to repress brain expansion by limiting neuronal progenitor production (Hill & Petersen 2015). However, it should also be noted that in the mammalian CNS, Sonic Hedgehog has known mitogenic roles, where its local production by differentiating or mature neurons signals back onto neural progenitors and adult neural stem cells to promote cell proliferation levels (Alvarez-Buylla & Ihrie 2014; Ihrie et al., 2011). Interestingly, this wasn’t the only parallel with the mammalian CNS, as Nkx2.1 and Arx are part of a transcription factor cascade within the early ventral telencephalon that specifies GABAergic and cholinergic interneurons (Butt et al., 2008; Vogt et al., 2014). I find it extremely interesting that across this huge evolutionary gap, these same transcription factors and this hedgehog signaling axis between mature neurons and stem cells have retained the same developmental functions in vastly different organisms. It also gives me reassurance about the importance of studying non-traditional model organisms, as the underlying basic biology is often conserved, so our findings from planarians, Hydra, Nematostella, Parhyale and other emerging lab models can be hugely informative for future studies on mammals and even humans.
References
Alvarez-Buylla A & Ihrie RA. (2014). Sonic hedgehog signaling in the postnatal brain. Seminars in Cell & Developmental Biology 33: 105-111. http://www.sciencedirect.com/science/article/pii/S1084952114001451
Butt SJ, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S & Fischell G. (2008). The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron 59: 722-732. http://www.sciencedirect.com/science/article/pii/S0896627308006302
Gurley KA, Rink JC & Sanchez Alvarado A. (2008). β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319: 323–327. http://science.sciencemag.org/content/319/5861/323
Hill EM & Petersen CP. (2015). Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142(24): 4217-29. http://dev.biologists.org/content/142/24/4217
Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kreigstein AR & Alvarez-Buylla A. (2011). Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron 71: 250-262. http://www.sciencedirect.com/science/article/pii/S0896627311004041
Morgan, T. H. (1898). Experimental studies of the regeneration of Planaria maculata. Arch. Entwm. Org. 7: 364–397. *This paper was subject of one of the Node’s Forgotten Classics posts*
Pallas, P. S. (1766). Miscellanea zoologica, quibus novae imprimis atque obscurae animalium species. Hagae Comitum, apud Pterum van Cleef, Holland.
Reddien PW, Bermange AL, Kicza AM & Sanchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134: 4043–4051. http://dev.biologists.org/content/134/22/4043
Reddien PW, Bermange AL, Murfitt KJ, Jennings JR & Sanchez Alvarado A. (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Developmental Cell 8: 635–649. http://www.sciencedirect.com/science/article/pii/S1534580705000924
Rink JC, Gurley KA, Elliott SA & Sanchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326: 1406–1410. http://science.sciencemag.org/content/326/5958/1406.long
Robb SM, Ross E & Sanchez Alvarado A. (2008). SmedGD: the Schmidtea mediterranea Genome Database. Nucleic Acids Res 36: D599–D606.
Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, Yang Z, Baraban SC & Rubenstein JL. (2014). Lhx6 directly regulates Arx and Cxcr7 to determine cortical interneuron fate and laminar position. Neuron 82: 350-364. http://www.cell.com/neuron/abstract/S0896-6273(14)00161-5


 (3 votes)
(3 votes)
This is awesome info! Much hope for more pieces of the puzzle! Kudos! To the Pearson lab!