BSDB Gurdon/The Company of Biologists 2019 Summer Studentship Report – Grace Blakeley
Posted by BSDB, on 15 January 2020
Established by the British Society for Developmental Biology in 2014, The Gurdon/The Company of Biologists Summer Studentship scheme provides financial support to allow highly motivated undergraduate students an opportunity to engage in practical research during their summer vacation. Each year, ten successful applicants spend eight weeks in the research laboratories of their choices, and the feedback we receive is outstanding. You can read accounts from previous years here. If you’re interested in applying or hosting a student in 2020, applications need to be in by the end of March.
Our second report from the class of 2019 comes from Grace Blakeley who studied spider segmentation in Alistair McGregor’s lab in Oxford Brookes University.
How to build a spider: investigating the role of Delta in posterior segmentation
Three animal phyla have segmented bodies – the vertebrate chordates, the annelid worms and the arthropods. To understand the evolution and development of these segmented bodies it is necessary to identify what mechanisms regulate segmentation and the similarities and differences of these mechanisms among phyla. Amongst arthropods there are two main different mechanisms for segmentation: long-germ arthropods, such as Drosophila melanogaster which develop their segments simultaneously; and short-germ arthropods, constituting the majority of arthropods, which add their posterior segments sequentially from a segment addition zone (SAZ). Under supervision from Dr Anna Schonauer in Professor Alistair McGregor’s lab, I aimed to investigate the regulation of posterior segmentation in arthropods by further studying the Delta-Notch signalling pathway in the spider Parasteatoda tepidariorum.
Spiders are a useful model for answering questions about the regulation and evolution of segmentation as they develop their prosomal (anterior) segments simultaneous like Drosophila but their opisthosomal (posterior) segments sequentially in a manner analogous vertebrate segmentation. It is understood that the addition of posterior segments relies on the formation of the SAZ, which develops at around stage 6 in spider embryos through dynamic Wnt and Delta-Notch signalling. These genes are subsequently also required for segment addition. Specifically, Delta appears to differentially regulate the posterior and anterior regions of the SAZ to maintain Wnt8 in the posterior SAZ but lower its expression in the anterior SAZ to facilitate the formation of nascent segments. However, little is known about the genes downstream of Delta that are involved in segment addition.
The gene hairy is thought to be involved in segment addition and it has been shown to have a similar oscillatory expression pattern to Delta leading to the hypothesis that it may be regulated by Delta as part of a gene regulatory network (GRN) that results in segment addition. Previous studies of the role of Delta in segment addition used parental RNA interference to knockdown this gene. However, the consequential loss of the SAZ resulting in a truncated germ band impedes investigation into any specific, localised downstream effects of loss of Delta. To be able to investigate the relationship between Delta and hairy I therefore aimed to use embryonic RNAi to knockdown Delta in subsets of posterior cells. This would allow embryonic development to progress as normal without truncating the germband, but create Delta knockdown clones allowing me to see within an embryo if hairy expression is different in cells with and without a knockdown of Delta. Thus I would be able to test whether hairy does act downstream of Delta and if so, might be able to infer what potential role it may have in segment addition.
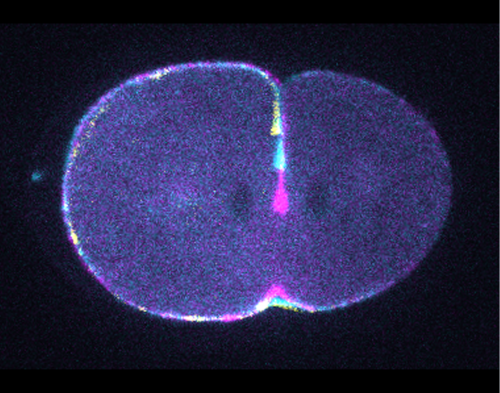
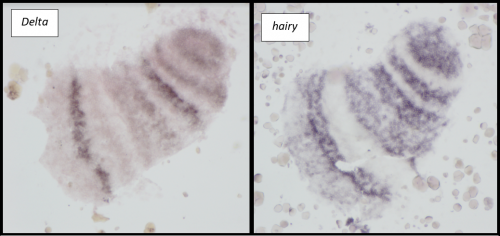
Microinjections of Delta double stranded RNA and biotin into single cells at embryonic stage 1F (when there are only 32 cells) were carried out with the aim of knocking down Delta and at the same time staining the clone of cells derived from the injected cell. The microinjection technique itself was technically demanding, requiring a lot of patience and practice and it took me two weeks before I was able to successfully inject a cell without it bursting. Once I had injected successfully into a single cell (Figure 1) I gained experience in the correct technique needed and I was then able to successfully inject about 15% of embryos in a cocoon (a cocoon containing approximately 200 embryos). The other main technique I used was in situ hybridisations (ISH), which used labelled RNA probes to bind to the hairy mRNA in all the cells in the embryo to show where hairy is being expressed. I carried out the ISH two days after the microinjections as this is when embryos develop their SAZ and begin to add their first segments. As a control, I stained embryos solely for Delta or hairy as this allowed me to see the wildtype expression of each (Figure 2). I also performed a double ISH for Delta and hairy which confirmed their overlapping expression in the posterior of the embryo.
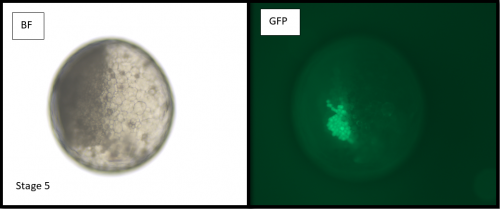
No blastodermal cell fate map exists for P. tepidariorum and as such it was never guaranteed that I would get clones in the posterior opisthosomal segments. Unfortunately, all my Delta knockdown clones occurred in the anterior prosomal segments so I was unable to draw conclusions about any Delta-hairy interaction in the SAZ. My anteriorly located clones however, do suggest an interaction between Delta and hairy in the prosomal segments. Here, I was able to detect downregulation of hairy expression in the subset of Delta knockdown cells. Most of our clones occur in a single prosomal segment, however in a stage 8 embryo, the clone spanned four developing prosomal segments, and showed downregulation of hairy across all of them. This suggests that Delta directs hairy in patterning of the anterior segments (data not shown).
Over the course of this project I learned many different things, most importantly that in science, patience and perseverance are key. I was surprised with how involved each technique was and quickly learned to take the advised protocol amendments suggested by my peers. Being part of a team all of whom supported me, helped me and questioned my work ultimately encouraged me to become a better researcher. The experience has confirmed my ambition to pursue a PhD and focused my interest on understanding the fundamental and complex molecular interactions that ultimately regulate and drive development.



 (2 votes)
(2 votes) (No Ratings Yet)
(No Ratings Yet)