A day in the life of a Platynereis dumerilii lab
Posted by Maggie Pruitt, on 25 June 2014
Hello! I am Maggie Pruitt, a postdoctoral researcher in the Department of Genetics, Development, and Cell Biology at Iowa State University (Ames, Iowa, USA – think middle America or fields upon fields). I work in Dr. Stephan Schneider’s evo-devo laboratory, and my work mostly focuses on studying components of the Wnt/β-catenin signaling pathway during early Platynereis dumerilii development.
The Schneider lab is part of the small, but growing, Platynereis community. Many of the Platynereis labs seem to study developmental biology or evolutionary developmental biology. While the Schneider lab is focusing on early development and the early gene regulatory networks present in Platynereis, other labs work on eye and brain development, gametogenesis, the molecular mechanisms of how the moon affects the life cycle of Platynereis and how neuropeptides control swimming depth of Platynereis larvae within the water column, to name a few.
Platynereis dumerilii as a model organism
Platynereis is a polychaete annelid and belongs to the clade Lophotrochozoa, which is the third but understudied branch of bilaterally symmetric animals (understudied compared to ecdysozoans like the nematode C. elegans, and deuterostomes like the vertebrate mouse). These worms can be collected in the wild from the coasts of the Mediterranean or coasts of Western Europe; however, they can also be easily cultured in a lab.
Our culture room. Notable features include boxes of adult or young worms, our artificial moonlight, and an injection microscope (yes, we can inject Platynereis embryos!).
As Platynereis is a marine worm, the worms need seawater. Filtered natural seawater (NSW) is by far the best for the worms. But, if your lab is not located close to a seawater source (like us in the middle of the USA) or you want to avoid steep costs for shipping NSW to your lab, you need to get a little creative. Artificial seawater (ASW) can be made using a salt called Instant Ocean. While this can be technically called seawater, the worms (or more specifically the embryos) do not really survive well in it. So…. since the Schneider lab is far from the sea and the worms do not like ASW, what do we use? This is where creativity comes in. Actually, it is where Dr. Dennis Lavrov’s sponge lab (also at Iowa State University) comes in. We provide the salt to make ASW to the Lavrov lab and their sponges. Then, once this seawater has been “conditioned” by the sponges (i.e. when the Lavrov lab needs to change the water in their tanks), we take their wastewater, filter it, sterilize it, and voila! Seawater that our embryos like, cleverly named DSW (Dennis seawater). We use 100% DSW for all of the fertilizations. Once the worms are juveniles or adults, we keep them in a 50/50 mixture of DSW and ASW.
The Schneider lab Platynereis culture is not huge, so optimal breeding success is key. One way to maximize this is to keep the culture on a fairly tight routine. The culture should be maintained at 18oC or 64.4oF, a standard for Platynereis labs around the globe, as the speed of early development is very temperature sensitive. In addition to constant temperature, it is good to have the worms on a regular feeding and water change schedule. Our young and adult worms receive minced organic spinach (only the best, pesticide-free spinach is good enough) on Mondays and ground fish food flakes on Fridays. The last incredibly important thing is to have tight control of the daily light and dark periods.
Sexual maturity and spawning in Platynereis dumerilii are synchronized by a lunar cycle. In the lab, the culture is on an artificial light cycle. The worms experience summer all year round, and so have 16 hours of light and 8 hours of total darkness each day. In addition, they are exposed to a 28-day night cycle in which three weeks of pitch-dark nights are followed by 7-days of full moon. Our high-tech recreation of this lunar cycle comes in the form of a $1.99 night light from Walmart, and a very reliable undergraduate researcher as a timer to turn the light on and off on the correct days. Sexually mature worms can be collected 3-6 days after our “moon” phase. Any disruption in this light cycle (exposing the worms to light during the dark phase), like leaving a digital clock on that has a bright backlight, has detrimental effects on the breeding success.
Sexual maturity can occur in worms as young as three months old, but sometimes we have “geriatric” 18-month-old worms finally maturing. In Platynereis, completed sexual maturity lasts one day/night, so an artificial cycle with artificial light allows for collective sexual maturity in the culture. Maturing worms are easily distinguished from non-mature worms by their change in appearance. Maturing worms have an empty gut and become more opaque. As the worms continue to mature, the females become yellow (lots of eggs) and the males have a cream anterior and a red posterior (lots of sperm and blood vessels).
Anterior halves of a female worm (left) and a male worm (right). The distinctive colors, yellow versus cream/red, are noticeable.
The awesome power of the Platynereis mating system
In the wild, Platynereis will swarm at dusk. However, in the lab we keep the sexually maturing male and female worms in separate bowls so the worms can last until the morning. Matings are easy to set-up; all you need is a glass bowl, some DSW, and a sexually mature male and female. After two worms are placed together in a bowl, they will exchange pheromones, which triggers their nuptial dancing and mating ritual:
The female is swimming in small circles, and the male is swimming in large circles. Pheromones are exchanged, the male first releases his cloud of sperm, and then the female releases her eggs (courtesy of Albrecht Fischer, University of Mainz, Germany).
Go ahead; watch the movie again. It is awesome and is partially responsible for getting me hooked on Platynereis. And at the end of this nuptial dance you are left with thousands of synchronously developing embryos and two dead worms. What a way to end your life, right? Perhaps before we pass judgment on how and when these worms die, I should give a bit more information on their life before the fatal spawning event.
Crash course to the life cycle of Platynereis
One of the many reasons why Platynereis is a good model system for developmental biology is that Platynereis develops rapidly. Platynereis begins as a 160μm egg and after instant fertilization of many eggs at once Platynereis embryos develop synchronously. In fact, the thousands of embryos from a single mating develop synchronously through the embryonic and larval stages. During early development, Platynereis embryos exhibit spiral cleavage. Spiral-cleaving embryos develop by a series of stereotyped asymmetric cell divisions that allows for the identification of individual cells by their positions and size. By 24h, the embryos have developed into a free-swimming larval form, a trochophore that swims freely within the water column, and after just 96h, Platynereis is a three-segmented juvenile worm that stays on the bottom of the dish. Segments are added throughout the lifetime of Platynereis at the tail end from a posterior growth zone, and Platynereis maintains a lifelong capability to grow and regenerate posterior segments after loss. The size of an adult worm varies tremendously, but on average they are about 35mm. During segment proliferation, the sexually immature worms live in self-spun silk tubes. The tubes are open at both ends and allow the worm to “check out” their environment, attack passing worms and prey, and eat their food. In their own world they are mostly holistic vegetarians that turn into fierce cannibalistic predators if the opportunity arises.
Here is one of approximately 90 boxes where we house our young and adult worms. In this box, you can see the many worms, each with their own silk tubes. If you look closely, you can even see one worm peeking out of its tube (right of center).
To sexually mature, Platynereis goes through a process called epitoky. Essentially, the whole body of the worm is irreversibly modified for reproduction, and the worm is transitioning from a sexually immature “atoke” form, to a sexually mature “epitoke” form. Some of the radical body modifications during this process include upgrades of motor and sensory organs, tune-ups of the muscular system for speedy swimming, resorbing of some muscles and all of the gut tissues, and the growing of thousands of gametes within their central body cavity. Indeed, the female and male worms are mostly reduced to growth chambers for gametes, bags of eggs and sperm, respectively. Once epitoky is complete, and the worm has reached its climax — one-day of sexual maturity, the worm will leave its silk tube home and swim into the pelagic zone (open water column). Here they will find like-minded partners (the good) or end up as a healthy protein shake for some higher ups in the food chain (the bad). After reproduction, the worms die. Maybe this death is still shocking to you, but the entire body of the worm changed for this one event…. the worm dies after completing a final task. From an evolutionary standpoint, the only task that matters.
A typical day in our laboratory
An ideal day in the lab is one in which everything is working correctly. Since this seems to be a mythical creature, I’ll stick to a typical day in our lab. Our typical days come in two flavors, depending on whether or not we have mature worms for fertilizations.
In the absence of sexually mature worms, a typical day in the lab consists of cloning, cloning, cloning, and maybe some more cloning. The goal of most of the cloning work is to make an in situ probe, and determine the expression pattern of the gene over the first 24h of development. The lab has an ever-growing list of interesting genes to clone, so this is a lab effort. I seem to spend most of my time working with all of the hard-earned embryos I collected in previous weeks (it can be hard to share embryos from late night fixations or embryos treated with pharmacological inhibitors). My favorite, and most frequent, activity is performing in situs. Cloning a new gene can be quite satisfying, but not nearly as much as uncovering a beautiful expression pattern that perfectly fits your hypothesis.
In the presence of sexually mature worms, I can typically be found running between our molecular lab (located on the 5th floor) and our culture room (located on the 7th floor). Maybe I’m being silly, but I feel extremely lazy taking the elevator up two flights of stairs. Since we only have sexually mature worms for less than two weeks each month, I try to take advantage of these days. Some of the fertilizations need to be saved to propagate the culture, but I try to use the rest. So, these days begin with a short run up to the culture room to set-up fertilizations. I’ll try to have a plan-of-action for the day, like how I plan to use the embryos, but sometimes, the worms just do not cooperate. Females weren’t feeling the males, all males and no females, etc. Pending total failure with the fertilizations, the embryos will be 1) used for microinjection or a pharmacological inhibitor experiment, 2) fixed for in situ hybridization or immunohistochemistry, or 3) used for RNA isolation. These days can get pretty hectic, but being busy means you and the lab will be set, sample-wise, for the weeks without the matures. It is only when slippery fingers make an appearance and multiple tubes of embryos or entire plates of treated embryos are dropped that these days are the worst. When this happens (yes, it happens), I usually get a coffee, then finish the day pretending the lab is in the absence of matures…. until the next morning.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.





 (8 votes)
(8 votes)
 In zebrafish, the circadian clock, which is the internal timekeeper that coordinates multiple cellular, physiological and behavioural processes with the external rhythmic environment, begins cycling very early in development. However, the functional relevance for embryonic and larval development of these early circadian oscillations is unclear. Here (p.
In zebrafish, the circadian clock, which is the internal timekeeper that coordinates multiple cellular, physiological and behavioural processes with the external rhythmic environment, begins cycling very early in development. However, the functional relevance for embryonic and larval development of these early circadian oscillations is unclear. Here (p.  A key question in developmental biology is how different tissues maintain proportional growth during development. A striking example of this is the tiling of sensory dendrites across the body wall of theDrosophila larva: during early larval life, the neuronal dendrites extend to cover the entire body wall, without overlapping. As the larva grows further, tiling is maintained – meaning that the dendrites and the overlying epithelium grow proportionally (dendrite-substrate coupling). On p.
A key question in developmental biology is how different tissues maintain proportional growth during development. A striking example of this is the tiling of sensory dendrites across the body wall of theDrosophila larva: during early larval life, the neuronal dendrites extend to cover the entire body wall, without overlapping. As the larva grows further, tiling is maintained – meaning that the dendrites and the overlying epithelium grow proportionally (dendrite-substrate coupling). On p.  The shoot apical meristem (SAM) is the growing tip of the plant stem, from which a population of pluripotent stem cells generates all above-ground organs. The SAM is organised both in a central-to-peripheral manner, with the central zone containing the stem cells while their progeny differentiate in the peripheral zone, and in outer-to-inner layers that generate different cell types. These different zones and layers of the SAM are presumably defined and regulated by distinct (if overlapping) gene regulatory networks, and G. Venugopala Reddy and co-workers (p.
The shoot apical meristem (SAM) is the growing tip of the plant stem, from which a population of pluripotent stem cells generates all above-ground organs. The SAM is organised both in a central-to-peripheral manner, with the central zone containing the stem cells while their progeny differentiate in the peripheral zone, and in outer-to-inner layers that generate different cell types. These different zones and layers of the SAM are presumably defined and regulated by distinct (if overlapping) gene regulatory networks, and G. Venugopala Reddy and co-workers (p.  DNA and histone methylation patterns correlate with – and define – transcriptional activity of the genome. In particular, DNA hypomethylation is associated with active chromatin and generally thought to be permissive for gene transcription. However, this rule is not globally applicable, and Shinichi Morishita, Hiroyuki Takeda and colleagues (p.
DNA and histone methylation patterns correlate with – and define – transcriptional activity of the genome. In particular, DNA hypomethylation is associated with active chromatin and generally thought to be permissive for gene transcription. However, this rule is not globally applicable, and Shinichi Morishita, Hiroyuki Takeda and colleagues (p. 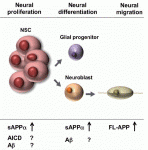 Interest in the amyloid precursor protein (APP) has increased in recent years due to its involvement in Alzheimer’s disease. Understanding the basic biology of APP and its physiological role during development thus will provide a better comprehension of Alzheimer’s disease.
Interest in the amyloid precursor protein (APP) has increased in recent years due to its involvement in Alzheimer’s disease. Understanding the basic biology of APP and its physiological role during development thus will provide a better comprehension of Alzheimer’s disease. 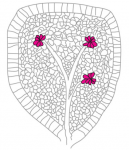 Multicellular rosettes have recently been appreciated as important cellular intermediates that are observed during the formation of diverse organ systems. Here, Nechiporuk and colleagues review recent studies of the genetic regulation and cellular transitions involved in rosette formation. They discuss and compare specific models for rosette formation and highlight outstanding questions in the field. See the Review on p.
Multicellular rosettes have recently been appreciated as important cellular intermediates that are observed during the formation of diverse organ systems. Here, Nechiporuk and colleagues review recent studies of the genetic regulation and cellular transitions involved in rosette formation. They discuss and compare specific models for rosette formation and highlight outstanding questions in the field. See the Review on p. 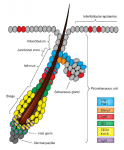 The epidermis is an integral part of our largest organ, the skin, and protects us against the hostile environment. Here, Jensen and co-workers discuss stem cell behaviour during normal tissue homeostasis, regeneration and disease within the pilosebaceous unit, an integral structure of the epidermis that is responsible for hair growth and lubrication of the epithelium. See the Review on p.
The epidermis is an integral part of our largest organ, the skin, and protects us against the hostile environment. Here, Jensen and co-workers discuss stem cell behaviour during normal tissue homeostasis, regeneration and disease within the pilosebaceous unit, an integral structure of the epidermis that is responsible for hair growth and lubrication of the epithelium. See the Review on p.  (No Ratings Yet)
(No Ratings Yet)