Pairing of segmentation clock genes results in robust pattern formation
Posted by Ertugrul Ozbudak, on 4 February 2021
By Oriana Q. H. Zinani, Kemal Keseroğlu, Ahmet Ay & Ertuğrul M. Özbudak:
Gene expression is an inevitably stochastic process (Ozbudak et al., 2002). In contrast, embryonic development and homeostasis require cells to coordinate the spatiotemporal expression of large sets of genes. Many mechanisms are known to orchestrate this coordination, such as operons, bidirectional promoters, enhancer sharing, 3-D DNA looping, topologically associated domains, transcription factories or hubs, and shared upstream regulators.
In metazoans, pairs of co-expressed genes often reside in the same chromosomal neighborhood, with gene pairs representing 10 to 50% of all genes, depending on the species (Adachi and Lieber, 2002; Arnone et al., 2012). Although many paired genes encode for essential housekeeping proteins, some encode for signaling regulators and transcription factors (such as CYP26A1–CYP26C1, ETS1–FLI1, MRF4–MYF5, MESP1–MESP2, SIX1–SIX4–SIX6, and STAT1–STAT4), which have transcription start sites that are 9–516 kb apart. Because various other mechanisms can ensure correlated gene expression, the selective advantage of maintaining adjacent gene pairs remains unknown.
To address this question, in Zinani et al, we used two linked zebrafish segmentation clock genes, her1–her7, as the testbed. The subdivision of the anterior–posterior axis into a fixed number of somites is a landmark example of how coordinated gene expression patterns the vertebrate embryo. During somitogenesis, groups of cells synchronously commit to segmentation in a notably short time frame. The pace of segmentation is set by the period of an oscillator, the segmentation clock, in cells of the unsegmented presomitic mesoderm (PSM). Oscillatory expression of the Hes or her clock genes is conserved in vertebrates; disruptions of their oscillations lead to vertebral segmentation defects (i.e., congenital scoliosis in humans). Approximately every 30 min in zebrafish, around 200 cells bud from the PSM to form a new somite. Segmentation is carried out for a species-specific number of cycles (33 in zebrafish). Her1 and Her7 are basic helix-loop-helix (bHLH) proteins that dimerize to bind DNA. The zebrafish segmentation clock relies on a transcriptional negative-feedback loop in which Her7-Hes6 hetero- or Her1-Her1 homodimers repress transcription of her1 and her7 (Ay et al., 2013; Giudicelli et al., 2007; Schroter et al., 2012). In zebrafish, two paired clock genes (her1 and her7) are separated by a 12-kb regulatory intervening sequence. her1 and her7 have similar transcriptional time delays (Hanisch et al., 2013) and RNA half-lives (Giudicelli et al., 2007); therefore, the transcription of her1 and her7 is mainly concomitant in the tissue. To achieve the rapid tempo and reproducible precision of segmentation, the transcription of her1 and her7 should be tightly coordinated.
Chromosomal adjacency was previously shown to cause correlated expression of synthetic reporters (Becskei et al., 2005; Fukaya et al., 2016; Raj et al., 2006). To test the role of gene pairing on transcriptional co-firing, we detected nascent transcription loci in the nucleus of single cells with single-molecule fluorescence in situ hybridization (smFISH) (Keskin et al., 2018; Zinani et al., 2021). We found that the probability of transcriptional co-firing of paired her1 and her7 genes on the same chromosome is significantly higher than the two unpaired her1 genes on homolog chromosomes. This finding demonstrated that gene pairing augments correlated transcription of the two clock genes by triggering transcriptional co-firing.
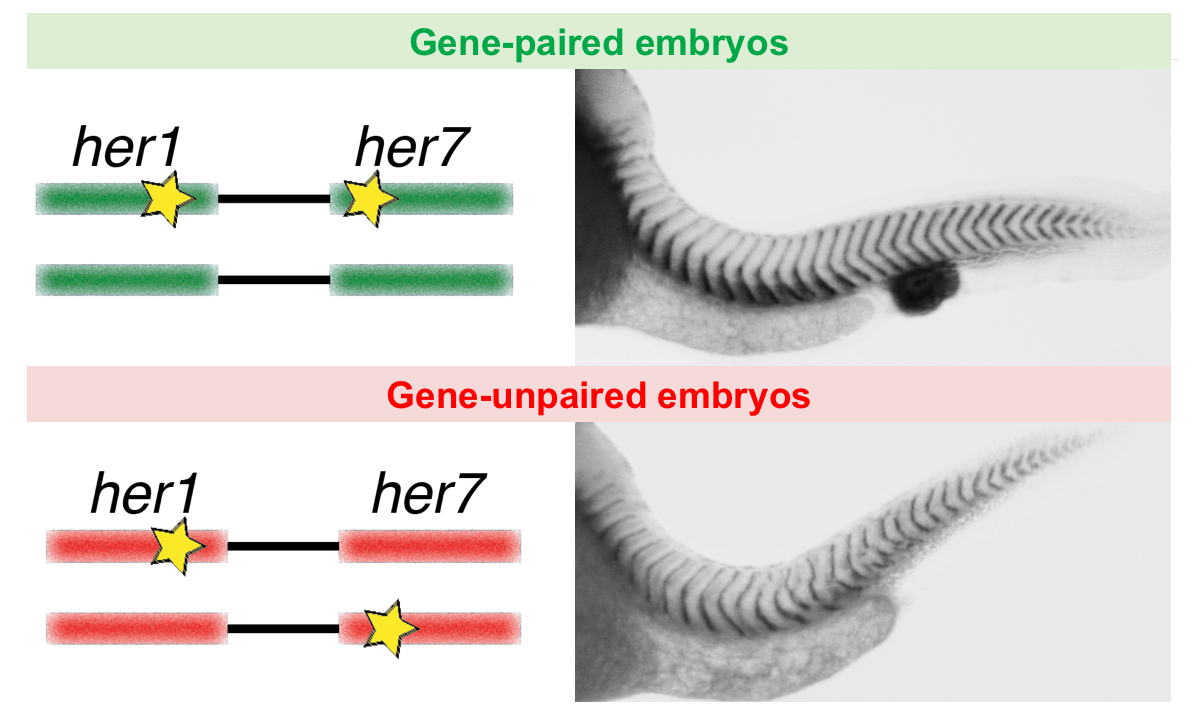
Co-firing of two paired clock genes could be advantageous for somite segmentation as it would coordinate transcript levels. To test this hypothesis, we generated cis and trans double heterozygous embryos by using CRISPR/Cas9 (Figure). The cis heterozygous mutants carried two mutant genes in one chromosome and two wild-type genes on the homologous chromosome (Figure, top row). In contrast, the trans mutants carried a mutant her1 gene adjacent to a wild-type her7 gene on one chromosome, and a mutant her7 gene adjacent to a wild-type her1 gene on the other chromosome (Figure, bottom row). Hence, fish that are compound heterozygous for these alleles will have the same functional gene dose as the previously described double heterozygous embryos with paired her1 and her7 genes (Figure, left column). We next raised the gene-paired and gene-unpaired embryos at 21.5 °C, where wild-type embryos successfully form somites. Gene-unpaired embryos had reduced success in somite segmentation as compared to gene-paired embryos (Figure, right column). These results revealed that maintaining paired genes in the genome is beneficial for successful pattern formation during embryonic development.
We next investigated the mechanism by which gene pairing is beneficial for clock oscillations. Although negative-feedback loops are widespread in gene regulatory networks, they usually do not give rise to oscillations, but instead act as a rheostat to tightly maintain gene expression around a steady-state. For a negative-feedback loop to generate oscillations, several important criteria need to be satisfied. Previous studies revealed the importance of time delays and short RNA or protein half-lives to generate sustained oscillations. However, another important criterion needed to generate oscillations is that the rate of transcription needs to be high enough to push the system into an unstable steady-state, establishing a limit cycle (Lewis, 2003; Novak and Tyson, 2008). We found that when genes are paired on the same chromosome, co-transcription happens more frequently. We hypothesized that co-firing of transcription results in a high rate of RNA production, and thereby overshoots the limit-cycle threshold. Simulations of our stochastic model agreed with our hypothesis.
To assess the function of gene pairing in the segmentation clock in real-time, we imaged transgenic Tg(her1:her1-Venus) (Delaune et al., 2012) zebrafish embryos along the entire PSM. We quantified the amplitude of oscillations in the next presumptive somites for 11 somite cycles. We found that the average amplitude was decreased in gene-unpaired embryos compared to gene-paired embryos. The amplitudes of oscillations preceding disrupted boundaries were significantly lower than the ones preceding the successful ones in a given genetic background.
Our results demonstrate that the prevention of gene pairing disrupts oscillations and segmentation in zebrafish embryos. We predict that gene pairing is similarly advantageous in other biological systems, and our findings could inspire engineering of precise synthetic clocks in embryos and organoids.
References:


 (6 votes)
(6 votes)